PGLYRP1-mediated intracellular peptidoglycan detection promotes intestinal mucosal protection
- PMID: 39984444
- PMCID: PMC11845746
- DOI: 10.1038/s41467-025-57126-9
PGLYRP1-mediated intracellular peptidoglycan detection promotes intestinal mucosal protection
Abstract
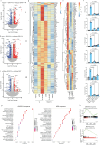
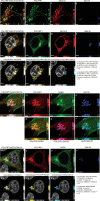
Peptidoglycan recognition proteins (PGLYRPs) are implicated in the control of the intestinal microbiota; however, molecular requirements for peptidoglycan (PGN) binding and receptor signaling mechanisms remain poorly understood. Here we show that PGLYRP1 is a receptor for the disaccharide motif of lysine N-acetylglucosamine N-acetylmuramic tripeptide (GMTriP-K). PGLYRP1 is required for innate immune activation by GMTriP-K but not muramyl dipeptide (MDP). In macrophages, intracellular PGLYRP1 complexes with NOD2 and GEF-H1, both of which are required for GMTriP-K-regulated gene expression. PGLYRP1 localizes to the endoplasmic reticulum and interacts at the Golgi with NOD2 upon GMTriP-K stimulation. PGLYRP1 and dependent gene expression signatures are induced in both mouse intestinal inflammation and human ulcerative colitis. Importantly, PGLYRP1 activation by GMTriP-K can result in the protection of mice from TNBS-induced colitis. Mammalian PGLYRPs can function as intracellular pattern recognition receptors for the control of host defense responses in the intestine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






Update of
-
PGLYRP-1 mediated intracellular peptidoglycan detection promotes mucosal protection.Res Sq [Preprint]. 2024 Oct 14:rs.3.rs-5118704. doi: 10.21203/rs.3.rs-5118704/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Feb 21;16(1):1864. doi: 10.1038/s41467-025-57126-9. PMID: 39483916 Free PMC article. Updated. Preprint.
References
-
- Dziarski, R. & Gupta, D. Review: mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun.16, 168–174 (2010). - PubMed
-
- Dziarski, R., Platt, K. A., Gelius, E., Steiner, H. & Gupta, D. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood102, 689–697 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

