Longitudinal evaluation of circulating tumor DNA in patients undergoing neoadjuvant therapy for early breast cancer using a tumor-informed assay
- PMID: 39984446
- PMCID: PMC11845481
- DOI: 10.1038/s41467-025-56658-4
Longitudinal evaluation of circulating tumor DNA in patients undergoing neoadjuvant therapy for early breast cancer using a tumor-informed assay
Abstract
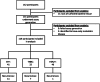
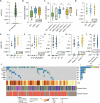
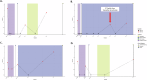
Circulating tumor DNA (ctDNA) is an emerging biomarker for the treatment of early breast cancer (EBC). We sought to evaluate a highly sensitive tumor-informed ctDNA assay in a real-world cohort of patients receiving neoadjuvant therapy (NAT) to assess clinical validity and explore prognostic outcomes. ctDNA is detected in 77.2% (88/114) of participants at baseline, with 18/88 (20.5%) having a baseline estimated variant allele frequency (eVAF) of <0.01%. Persistent detection of ctDNA, measured midway through NAT (mid-NAT), is associated with disease recurrence in all participants, reaching statistical significance in those with HER2-negative disease. Stratified analyses demonstrate that ctDNA detected mid-NAT enhances the prognostic accuracy of the residual cancer burden (RCB) score for disease recurrence. Postoperative or follow-up detection of ctDNA demonstrates a 100% positive predictive value for disease recurrence, with a median lead time of 374 days (range: 13-1010 days). These data suggest that assays with high analytical sensitivity may improve baseline ctDNA detection in patients with EBC. The ability to replicate the prognostic association of ctDNA dynamics in a real-world cohort supports further investigation. Prospective trials incorporating ctDNA testing are warranted to assess and develop the clinical utility of ctDNA-guided treatment strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.J.E, P.E., J.F.A., S.M., E.G.A., A.D., Z.V., N.M., E.S., C.Y., and H.K.B. report no competing interests. E.A. reports receiving honoraria from Sandoz and Seagen and consulting fees from AstraZeneca and Novartis. M.B.N. reports speaker honoraria and consulting fees from Novartis and Exact Sciences, outside the scope of this submitted work. N.C., R.V., and C.P. were employees of NeoGenomics at the time of data generation and manuscript preparation. P.L.B. reports research funding (to the institution) from Amgen, AstraZeneca, Bayer, Bicara, Bristol Myers Squibb, Genentech/Roche, Gilead, GlaxoSmithKline, Lilly/Loxo, Medicenna, Merck, Nektar, Novartis, PTC Therapeutics, Sanofi, SeaGen, Servier, SignalChem Life Sciences, Takeda, and Zymeworks. He also reports uncompensated honoraria/consultancy with Zymeworks, Lilly, Seattle Genetics, Merck, Amgen, Gilead, Janssen, and Repare. L.L.S. serves on scientific advisory boards for Merck, Pfizer, AstraZeneca, Roche, GlaxoSmithKline, Voronoi, Arvinas, Navire, Relay, Daiichi Sankyo, Amgen, Marengo, Medicenna, Tubulis, LTZ Therapeutics, Pangea, and Break Through Cancer. She also reports clinical trial support (to the institution) from Novartis, Bristol Myers Squibb, Pfizer, Boehringer Ingelheim, GlaxoSmithKline, Roche/Genentech, AstraZeneca, Merck, Celgene, Astellas, Bayer, AbbVie, Amgen, Symphogen, Mirati, BioNTech, 23andMe, and EMD Serono. Additionally, her spouse holds stock ownership in Agios and has leadership roles at Treadwell Therapeutics. D.W.C. reports consultancy and advisory relationships with AstraZeneca, Daiichi Sankyo, GenomeRx, Gilead, GlaxoSmithKline, NeoGenomics, Lilly, Merck, Novartis, Pfizer, Roche and SAGA; research funding (to the institution) from AstraZeneca, Guardant Health, Grail, Gilead, GlaxoSmithKline, NeoGenomics, Knight, Merck, Pfizer, ProteinQure, and Roche.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

