TGM6 is a helminth secretory product that mimics TGF-β binding to TGFBR2 to antagonize signaling in fibroblasts
- PMID: 39984487
- PMCID: PMC11845725
- DOI: 10.1038/s41467-025-56954-z
TGM6 is a helminth secretory product that mimics TGF-β binding to TGFBR2 to antagonize signaling in fibroblasts
Abstract
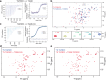
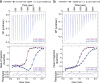
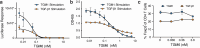
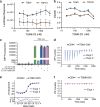
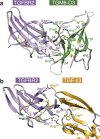
TGM6 is a natural antagonist of mammalian TGF-β signaling produced by the murine helminth parasite Heligmosomoides polygyrus. It differs from the previously described agonist, TGM1 (TGF-β Mimic-1), in that it lacks domains 1/2 that bind TGFBR1. It nonetheless retains TGFBR2 binding through domain 3 and potently inhibits TGF-β signaling in fibroblasts and epithelial cells, but does not inhibit TGF-β signaling in T cells, consistent with divergent domains 4/5 and an altered co-receptor binding preference. The crystal structure of TGM6 bound to TGFBR2 reveals an interface remarkably similar to that of TGF-β with TGFBR2. Thus, TGM6 has adapted its structure to mimic TGF-β, while engaging a distinct co-receptor to direct antagonism to fibroblasts and epithelial cells. The co-expression of TGM6, along with immunosuppressive TGMs that activate the TGF-β pathway, may minimize fibrotic damage to the host as the parasite progresses through its life cycle from the intestinal lumen to submucosa and back again. The co-receptor-dependent targeting of TGFBR2 by the parasite provides a template for the development of therapies for targeting the cancer- and fibrosis-promoting activities of the TGF-βs in humans.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.H. is a member of the scientific advisory board for TCGFB and is a licensee of intellectual property to Kalivir Immunotherapeutics. The remaining authors declare no competing interests.
Figures



Update of
-
TGM6, a helminth secretory product, mimics TGF-β binding to TβRII to antagonize TGF-β signaling in fibroblasts.bioRxiv [Preprint]. 2023 Dec 23:2023.12.22.573140. doi: 10.1101/2023.12.22.573140. bioRxiv. 2023. Update in: Nat Commun. 2025 Feb 21;16(1):1847. doi: 10.1038/s41467-025-56954-z. PMID: 38187573 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

