STIM1-mediated NFAT signaling synergizes with STAT1 to control T-bet expression and TH1 differentiation
- PMID: 39984734
- PMCID: PMC12121662
- DOI: 10.1038/s41590-025-02089-8
STIM1-mediated NFAT signaling synergizes with STAT1 to control T-bet expression and TH1 differentiation
Abstract
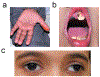
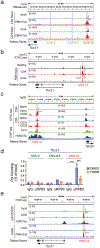
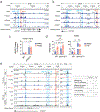
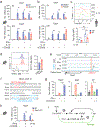
Stromal interaction molecule 1 (STIM1) is critical for store-operated Ca2+ entry (SOCE) and T cell activation. T helper 1 (TH1) cells, which express T-bet (encoded by TBX21), mediate immunity to intracellular pathogens. Although SOCE is known to regulate other TH lineages, its role in Th1 differentiation remains unclear. Here, we report a patient with an intronic loss-of-function mutation in STIM1, which abolishes SOCE and causes immunodeficiency. We demonstrate that SOCE promotes nuclear factor of activated T cells (NFAT) binding to conserved noncoding sequence (CNS)-12 in the TBX21 enhancer and enables NFAT to synergize with STAT1 to mediate TBX21 expression. While SOCE-deficient CD4+ T cells have reduced expression of TBX21 in the absence of interleukin-12 (IL-12), their expression of IL-12 receptors β1 and β2 is increased, sensitizing them to IL-12 signaling and allowing IL-12 to rescue T-bet expression. Our study reveals that the STIM1-SOCE-NFAT signaling axis is essential for the differentiation of Th1 cells depending on the cytokine milieu.
© 2025. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: S.F. is a scientific cofounder and consultant of Calcimedica, and an inventor on a patent Regulators of NFAT (WO/2007/081804) related to this paper. The other authors declare no competing interests.
Figures










References
-
- Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000). - PubMed
METHODS-ONLY REFERENCES
-
- Tsvilovskyy V, et al. Deletion of Orai2 augments endogenous CRAC currents and degranulation in mast cells leading to enhanced anaphylaxis. Cell Calcium 71, 24–33 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- R01 AI130143/AI/NIAID NIH HHS/United States
- BMRP program/Qatar Foundation (Qatar Foundation for Education, Science and Community Development)
- AI164803/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- F30 AI164803/AI/NIAID NIH HHS/United States
- AI130143/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

