Different Ras isoforms regulate synaptic plasticity in opposite directions
- PMID: 39984756
- PMCID: PMC11961722
- DOI: 10.1038/s44318-025-00390-8
Different Ras isoforms regulate synaptic plasticity in opposite directions
Abstract
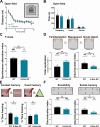
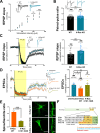
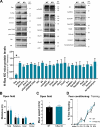
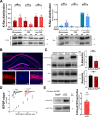
The small GTPase Ras is an intracellular signaling hub required for long-term potentiation (LTP) in the hippocampus and for memory formation. Genetic alterations in Ras signaling (i.e., RASopathies) are linked to cognitive disorders in humans. However, it remains unclear how Ras controls synaptic plasticity, and whether different Ras isoforms play overlapping or distinct roles in neurons. Using genetically modified mice, we show here that H-Ras (the most abundant isoform in the brain) does not promote LTP, but instead long-term depression mediated by metabotropic glutamate receptors (mGluR-LTD). Mechanistically, H-Ras is activated locally in spines during mGluR-LTD via c-Src, and is required to trigger Erk activation and de novo protein synthesis. Furthermore, H-Ras deletion impairs object recognition as well as social and spatial memory. Conversely, K-Ras is the isoform specifically required for LTP. This functional specialization correlates with a differential synaptic distribution of the two isoforms H-Ras and K-Ras, which may have important implications for RASopathies and cognitive function.
Keywords: Hippocampus; Memory; RASopathies; Spine; mGluR.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures







References
-
- Barbacid M (1987) ras genes. Annu Rev Biochem 56:779–827 - PubMed
MeSH terms
Substances
Grants and funding
- PID2022-136932OB-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- RYC2021-031395-I/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PID2020-117651RB/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PDC2021-120815-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PID2020-119358GB-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

