Contribution of germline and somatic mutations to risk of neuromyelitis optica spectrum disorder
- PMID: 39986280
- PMCID: PMC11960548
- DOI: 10.1016/j.xgen.2025.100776
Contribution of germline and somatic mutations to risk of neuromyelitis optica spectrum disorder
Abstract
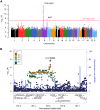
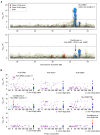
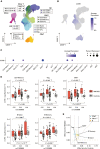
Neuromyelitis optica spectrum disorder (NMOSD) is a rare autoimmune disease characterized by optic neuritis and transverse myelitis, with an unclear genetic background. A genome-wide meta-analysis of NMOSD in Japanese individuals (240 patients and 50,578 controls) identified significant associations with the major histocompatibility complex region and a common variant close to CCR6 (rs12193698; p = 1.8 × 10-8, odds ratio [OR] = 1.73). In single-cell RNA sequencing (scRNA-seq) analysis (25 patients and 101 controls), the CCR6 risk variant showed disease-specific expression quantitative trait loci effects in CD4+ T (CD4T) cell subsets. Furthermore, we detected somatic mosaic chromosomal alterations (mCAs) in various autoimmune diseases and found that mCAs increase the risk of NMOSD (OR = 3.37 for copy number alteration). In scRNA-seq data, CD4T cells with 21q loss, a recurrently observed somatic event in NMOSD, showed dysregulation of type I interferon-related genes. Our integrated study identified novel germline and somatic mutations associated with NMOSD pathogenesis.
Keywords: clonal hematopoiesis; eQTL; genome-wide association study; human leukocyte antigen; mosaic chromosomal alterations; neuromyelitis optica spectrum disorder; scRNA-seq; single-cell RNA sequencing; somatic mutation.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Lennon V.A., Wingerchuk D.M., Kryzer T.J., Pittock S.J., Lucchinetti C.F., Fujihara K., Nakashima I., Weinshenker B.G. A serum autoantibody marker of neuromyelitis optica: distinction from multiple sclerosis. Lancet. 2004;364:2106–2112. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

