Integration of single-cell and bulk RNA sequencing to identify a distinct tumor stem cells and construct a novel prognostic signature for evaluating prognosis and immunotherapy in LUAD
- PMID: 39987127
- PMCID: PMC11847374
- DOI: 10.1186/s12967-025-06243-6
Integration of single-cell and bulk RNA sequencing to identify a distinct tumor stem cells and construct a novel prognostic signature for evaluating prognosis and immunotherapy in LUAD
Abstract
Background: Cancer stem cells (CSCs) are crucial for lung adenocarcinoma (LUAD). This study investigates tumor stem cell gene signatures in LUAD using single-cell RNA sequencing (scRNA-seq) and bulk RNA sequencing (RNA-seq), aiming to develop a prognostic tumor stem cell marker signature (TSCMS) model.
Methods: LUAD scRNA-seq and RNA-seq data were analyzed. CytoTRACE software quantified the stemness score of tumor-derived epithelial cell clusters. Gene Set Variation Analysis (GSVA) identified potential biological functions in different clusters. The TSCMS model was constructed using Lasso-Cox regression, and its prognostic value was assessed through Kaplan-Meier, Cox regression, and receiver-operating characteristic (ROC) curve analyses. Immune infiltration was evaluated using the Cibersortx algorithm, and drug response prediction was performed using the pRRophetic package. TAF10 functional investigations in LUAD cells involved bioinformatics analysis, qRT-PCR, Western blot, immunohistochemistry, and assays for cell proliferation.
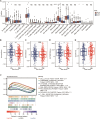
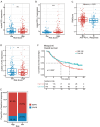
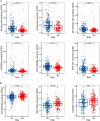
Results: Seven distinct cell clusters were identified by CytoTRACE, with epithelial cell cluster 1 (Epi_C1) showing the highest stemness potential. The TSCMS model included 49 tumor stemness-related genes; high-risk patients exhibited lower immune and ESTIMATE scores and increased tumor purity. Significant differences in immune landscapes and chemotherapy sensitivity were observed between risk groups. TAF10 positively correlated with RNA expression-based stemness scores in various tumors, including LUAD. It was over-expressed in LUAD cell lines and clinical tumor tissues, with high expression linked to poor prognosis. Silencing TAF10 inhibited LUAD cell proliferation and tumor sphere formation.
Conclusions: This study demonstrates the TSCMS model's prognostic value in LUAD, reveals insights into immune infiltration and therapeutic response, and identifies TAF10 as a potential therapeutic target.
Keywords: Gene signature; LUAD; Prognostic model; Single-cell RNA; Tumor stem cell.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The protocol and all procedures involving human samples in this study were reviewed and approved by the Institutional Review Board (IRB) of Zhongshan City People's Hospital (approval number: 2024–116). Informed patient consent was obtained from all individuals whose paraffin-embedded tissues were used in this study. All research procedures were conducted in compliance with relevant ethical guidelines and local laws. Consent for publication: Not appliable. Competing interests: The authors declare no conflict of interest.
Figures




References
-
- Suva ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell. 2019;75:7–12. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

