Interpretable machine learning to evaluate relationships between DAO/DAOA (pLG72) protein data and features in clinical assessments, functional outcome, and cognitive function in schizophrenia patients
- PMID: 39987274
- PMCID: PMC11846841
- DOI: 10.1038/s41537-024-00548-z
Interpretable machine learning to evaluate relationships between DAO/DAOA (pLG72) protein data and features in clinical assessments, functional outcome, and cognitive function in schizophrenia patients
Abstract
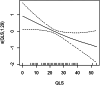
Machine learning has been proposed to utilize D-amino acid oxidase (DAO) and DAO activator (DAOA [or pLG72]) protein levels to ascertain disease status in schizophrenia. However, it remains unclear whether machine learning can effectively evaluate clinical features in relation to DAO and DAOA in schizophrenia patients. We employed an interpretable machine learning (IML) framework including linear regression, least absolute shrinkage and selection operator (Lasso) models, and generalized additive models (GAMs) to analyze DAO/DAOA levels using 380 Taiwanese schizophrenia patients. Additionally, we incorporated 27 parameters encompassing demographic variables, clinical assessments, functional outcomes, and cognitive function as features. The IML framework facilitated linear and non-linear relationships between features and DAO/DAOA. DAO levels demonstrated significant associations with the 17-item Hamilton Depression Rating Scale (HAMD17) based on linear regression. The Lasso model identified four features-HAMD17, age, working memory, and overall cognitive function (OCF)-and highlighted HAMD17 as the most significant feature, using DAO from chronically stable patients. Utilizing DAOA from acutely exacerbated patients, the Lasso model also identified four features-OCF, Scale for the Assessment of Negative Symptoms 20-item, quality of life scale (QLS), and category fluency-and emphasized OCF as the most significant feature. Furthermore, GAMs revealed a non-linear relationship between category fluency and DAO in chronically stable patients, as well as between QLS and DAOA in acutely exacerbated patients. The study suggests that an IML framework holds promise for assessing linear and non-linear relationships between DAO/DAOA and various features in clinical assessments, functional outcomes, and cognitive function in patients with schizophrenia.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Prediction Analysis for Transition to Schizophrenia in Individuals at Clinical High Risk for Psychosis: The Relationship of DAO, DAOA, and NRG1 Variants with Negative Symptoms and Cognitive Deficits.Front Psychiatry. 2017 Dec 20;8:292. doi: 10.3389/fpsyt.2017.00292. eCollection 2017. Front Psychiatry. 2017. PMID: 29326614 Free PMC article.
-
A systematic meta-analysis of the association of Neuregulin 1 (NRG1), D-amino acid oxidase (DAO), and DAO activator (DAOA)/G72 polymorphisms with schizophrenia.J Neural Transm (Vienna). 2018 Jan;125(1):89-102. doi: 10.1007/s00702-017-1782-z. Epub 2017 Sep 1. J Neural Transm (Vienna). 2018. PMID: 28864885
-
Evidence for association and epistasis at the DAOA/G30 and D-amino acid oxidase loci in an Irish schizophrenia sample.Am J Med Genet B Neuropsychiatr Genet. 2007 Oct 5;144B(7):949-53. doi: 10.1002/ajmg.b.30452. Am J Med Genet B Neuropsychiatr Genet. 2007. PMID: 17492767
-
The DAOA gene is associated with schizophrenia in the Taiwanese population.Psychiatry Res. 2017 Jun;252:201-207. doi: 10.1016/j.psychres.2017.03.013. Epub 2017 Mar 7. Psychiatry Res. 2017. PMID: 28285246
-
G72 primate-specific gene: a still enigmatic element in psychiatric disorders.Cell Mol Life Sci. 2016 May;73(10):2029-39. doi: 10.1007/s00018-016-2165-6. Epub 2016 Feb 25. Cell Mol Life Sci. 2016. PMID: 26914235 Free PMC article. Review.
References
-
- Katsanis, S. H., Javitt, G. & Hudson, K. Public health. A case study of personalized medicine. Science320, 53–54 (2008). - PubMed
-
- Snyderman, R. Personalized health care: from theory to practice. Biotechnol. J.7, 973–979 (2012). - PubMed
-
- Lane, H. Y., Tsai, G. E. & Lin, E. Assessing gene-gene interactions in pharmacogenomics. Mol. Diagn. Ther.16, 15–27 (2012). - PubMed
-
- Lin, E. & Chen, P. S. Pharmacogenomics with antidepressants in the STAR*D study. Pharmacogenomics9, 935–946 (2008). - PubMed
-
- Lin, E. & Lane, H. Y. Genome-wide association studies in pharmacogenomics of antidepressants. Pharmacogenomics16, 555–566 (2015). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

