STING mediates increased self-renewal and lineage skewing in DNMT3A-mutated hematopoietic stem/progenitor cells
- PMID: 39987368
- PMCID: PMC11976293
- DOI: 10.1038/s41375-025-02542-5
STING mediates increased self-renewal and lineage skewing in DNMT3A-mutated hematopoietic stem/progenitor cells
Abstract
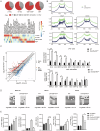
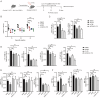
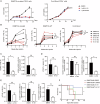
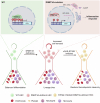
Somatic mutations in DNA methyltransferase 3 A (DNMT3A) are frequently observed in patients with hematological malignancies. Hematopoietic stem/progenitor cells (HSPCs) with mutated DNMT3A demonstrate increased self-renewal activity and skewed lineage differentiation. However, the molecular mechanisms underlying these changes remain largely unexplored. In this study, we show that Dnmt3a loss leads to the upregulation of endogenous retroviruses (ERVs) in HSPCs, subsequently activating the cGAS-STING pathway and triggering inflammatory responses in these cells. Both genetic and pharmacological inhibition of STING effectively corrects the increased self-renewal activity and differentiation skewing induced by Dnmt3a deficiency in mice. Notably, targeting STING showed inhibited acute myeloid leukemia (AML) development in a Dnmt3a-KO; Flt3-ITD AML model, comparable to AC220, an FDA-approved FLT3-ITD inhibitor. A patient-derived xenograft (PDX) model further demonstrated that targeting STING effectively alleviates the leukemic burden of DNMT3A-mutant AML. Collectively, our findings highlight a critical role for STING in hematopoietic disorders induced by DNMT3A mutations and propose STING as a potential therapeutic target for preventing the progression of DNMT3A mutation-associated leukemia.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: All animal experiments described in this study were conducted in accordance with the ethical guidelines of the Department of Laboratory Animal Science Fudan University, No. 202307021S. The primary leukemia cells were collected from patient bone marrow cells and followed the ethical guidelines of Huashan Hospital, No. KY2015-269.
Figures



References
-
- Okano M, Bell DW, Haber DA, Li E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell. 1999;99:247–57. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

