Interleukin-10 production by innate lymphoid cells restricts intestinal inflammation in mice
- PMID: 39988202
- PMCID: PMC12167137
- DOI: 10.1016/j.mucimm.2025.02.005
Interleukin-10 production by innate lymphoid cells restricts intestinal inflammation in mice
Abstract
Interleukin-10 (IL-10) is an immunomodulatory cytokine critical for intestinal immune homeostasis. IL-10 is produced by various immune cells but IL-10 receptor signaling in intestinal CX3CR1+ mononuclear phagocytes is necessary to prevent spontaneous colitis in mice. Here, we utilized fluorescent protein reporters and cell-specific targeting and found that Rorc-expressing innate lymphoid cells (ILCs) produce IL-10 in response to anti-CD40-mediated intestinal inflammation. Deletion of Il10 specifically in Rorc-expressing ILCs led to phenotypic changes in intestinal macrophages and exacerbated both innate and adaptive immune-mediated models of experimental colitis. The population of IL-10+ producing ILCs shared markers with both ILC2 and ILC3 with nearly all ILC3s being of the NCR+ subtype. Interestingly, Ccl26 was enriched in IL-10+ ILCs and was markedly reduced in IL-10-deficient ILC3s. Since CCL26 is a ligand for CX3CR1, we employed RNA in situ hybridization and observed increased numbers of ILCs in close proximity to Cx3cr1-expressing cells under inflammatory conditions. Finally, we generated transgenic RorctdTomato reporter mice that faithfully marked RORγt+ cells that could rescue disease pathology and aberrant macrophage phenotype following adoptive transfer into mice with selective Il10 deficiency in ILC3s. These results demonstrate that IL-10 production by a population of ILCs functions to promote immune homeostasis in the intestine possibly via direct effects on intestinal macrophages.
Keywords: CCL26; CX(3)CR1; IBD; IL-10; ILC3.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

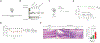

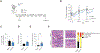
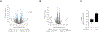


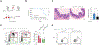
References
MeSH terms
Substances
Grants and funding
- IK2 BX004648/BX/BLRD VA/United States
- I01 CX002473/CX/CSRD VA/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 DK129552/DK/NIDDK NIH HHS/United States
- R01 HL163043/HL/NHLBI NIH HHS/United States
- R01 AI145930/AI/NIAID NIH HHS/United States
- P50 CA236733/CA/NCI NIH HHS/United States
- R01 AI171980/AI/NIAID NIH HHS/United States
- R03 DK123489/DK/NIDDK NIH HHS/United States
- R01 DK103831/DK/NIDDK NIH HHS/United States
- R01 DK128200/DK/NIDDK NIH HHS/United States
- S10 OD023475/OD/NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- I01 BX001426/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

