Transcription-replication conflicts drive R-loop-dependent nucleosome eviction and require DOT1L activity for transcription recovery
- PMID: 39988315
- PMCID: PMC11840560
- DOI: 10.1093/nar/gkaf109
Transcription-replication conflicts drive R-loop-dependent nucleosome eviction and require DOT1L activity for transcription recovery
Abstract
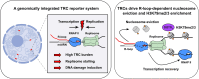
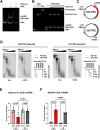
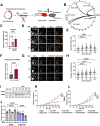
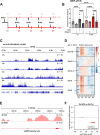
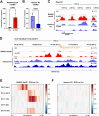
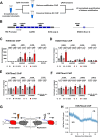
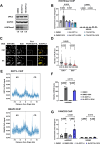
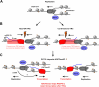
Progressing transcription and replication machineries profoundly impact their underlying chromatin template. Consequently, transcription-replication conflict (TRC) sites are vulnerable to chromatin and epigenome alterations, provoking genome instability. Here, we engineered an inducible TRC reporter system using a genome-integrated R-loop-prone sequence and characterized the dynamic changes of the local chromatin structure inflicted by TRCs, leading to reduced nucleosome occupancy and replication fork blockage. Strikingly, inducing a small number of TRCs on the genome results in a measurable global replication stress response. Furthermore, we find a TRC-dependent increase in H3K79 methylation specifically at the R-loop forming TRC site. Accordingly, inhibition of the H3K79 methyltransferase DOT1L leads to reduced transcriptional output and an exacerbated DNA damage response, suggesting that deposition of this mark is required for effective transcription recovery and resolution of TRCs. Our work shows the molecular dynamics and reveals a specific epigenetic modifier bookmarking TRC sites, relevant to cancer and other diseases.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

