OCT4 translationally promotes AKT signaling as an RNA-binding protein in stressed pluripotent stem cells
- PMID: 39988663
- PMCID: PMC11849194
- DOI: 10.1186/s13287-025-04229-1
OCT4 translationally promotes AKT signaling as an RNA-binding protein in stressed pluripotent stem cells
Abstract
Background: Despite numerous studies addressing the molecular mechanisms by which pluripotent stem cells (PSCs) maintain self-renewal and pluripotency under normal culture conditions, the fundamental question of how PSCs manage to survive stressful conditions remains largely unresolved. Post-transcriptional/translational regulation emerges to be vital for PSCs, but how PSCs coordinate and balance their survival and differentiation at translational level under extrinsic and intrinsic stress conditions is unclear.
Methods: The high-throughput sequencing of cross-linking immunoprecipitation cDNA library (HITS-CLIP) was employed to decipher the genome-wide OCT4-RNA interactome in human PSCs, a combined RNC-seq/RNA-seq analysis to assess the role of OCT4 in translational regulation of hypoxic PSCs, and an OCT4-protein interactome to search for OCT4 binding partners that regulate cap-independent translation initiation. By taking the Heterozygous Knocking In N-terminal Tags (HKINT) approach that specifically disrupts the 5'-UTR secondary structure and tagging its protein product of the mRNA from one allele while leaving that from the other allele intact, we examined the effect of disrupting the OCT4/5'-UTR interaction on translation of AKT1 mRNA.
Results: We revealed OCT4 as a bona fide RNA-binding protein (RBP) in human PSCs that bound to the 5'-UTR, 3'-UTR and CDS regions of mRNAs. Multiple known proteins participating in IRES-mediated translation initiation were detected in the OCT4-protein interactome, and a combined RNC-seq/RNA-seq analysis further confirmed a crucial role of OCT4 in translational regulation of PSCs in response to hypoxic stress. Remarkably, OCT4 bound to the GC-rich elements in the 5'-UTR of AKT1 and multiple PI3K/AKT-pathway-gene mRNAs, and promoted their translation initiation via IRES-mediated pathways under stress conditions. Specifically disrupting the AKT1 mRNA 5'-UTR structure and the OCT4/5'-UTR interaction by the HKINT approach significantly reduced the translation level of AKT1 that led to a higher susceptibility of PSCs to oxidative stress-induced apoptotic death and prioritized differentiation toward ectoderm and endoderm.
Conclusions: Our results reveal OCT4 as an anti-stress RBP for translational regulation that critically coordinates the survival and differentiation of PSCs in response to various stressors.
Keywords: AKT; OCT4; Oxidative stress; Pluripotent stem cells (PSCs); RNA-binding protein (RBP); Translation initiation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Human (h) ESC line H1 (SCSP-301) cells ( https://www.cellbank.org.cn/search-detail.php?id=483 ) and H9 (SCSP-302) cells ( https://www.cellbank.org.cn/search-detail.php?id=484 ) were acquired from the Cell Bank of the Chinese Academic of Sciences, Shanghai, China, under Material Transfer Agreements. The Cell Bank obtained prior ethical approval from WiCell Research Institute for the collection of hESC lines. The handling, storage and preparation of those cells was conducted following the protocols provided by the Cell Bank. All experiments using the obtained hESC lines in this study were approved by the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University, and were performed according to institutional guidelines. Title of the approved project: Endogenous OCT4 promoter-driven reporter system to study self-renewal of pluripotent and cancer stem cells. Name of the institutional approval committee or unit: Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University. Approval number: 2019–377. Date of approval: February 8, 2019. Consent for publication: Not applicable. Competing interest: The authors declare that they have no conflict of interest.
Figures

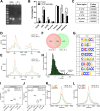




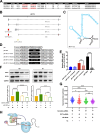

References
-
- Trounson A, DeWitt ND. Pluripotent stem cells progressing to the clinic. Nat Rev Mol Cell Biol. 2016;17(3):194–200. - PubMed
-
- Yu JSL, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143(17):3050–60. - PubMed
-
- Zhao H, Jin Y. Signaling networks in the control of pluripotency. Curr Opin Genet Dev. 2017;46:141–8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

