Chromobox protein homolog 7 suppresses the stem-like phenotype of glioblastoma cells by regulating the myosin heavy chain 9-NF-κB signaling pathway
- PMID: 39988672
- PMCID: PMC11847914
- DOI: 10.1038/s41420-025-02362-7
Chromobox protein homolog 7 suppresses the stem-like phenotype of glioblastoma cells by regulating the myosin heavy chain 9-NF-κB signaling pathway
Abstract
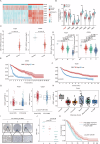
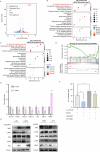
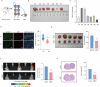
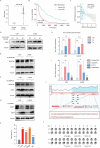
Cancer stem cells (CSCs) are significant factors in the treatment resistance and recurrence of glioblastoma. Chromobox protein homolog 7 (CBX7) can inhibit the progression of various tumors, but its impact on the stem cell-like properties of glioblastoma cells remains unclear. Clinically, low levels of CBX7 are associated with poor prognosis and increased distant metastasis in glioblastoma patients, and this low expression is caused by methylation of the CBX7 promoter. Our current research indicates that CBX7 plays a key role in suppressing the stem-like phenotype of glioblastoma. In this study, through bioinformatics analysis, we found that CBX7 is the most significantly downregulated member of the CBX family in glioblastoma and is closely associated with the stem-like phenotype of glioblastoma cells. We show that CBX7 promotes the degradation of myosin heavy chain 9 (MYH9) protein through the ubiquitin-proteasome pathway via the polycomb repressive complex 1 (PRC1) and suppresses the stem-like phenotype of glioblastoma cells by inhibiting the nuclear factor kappa-B (NF-κB) signaling pathway. Furthermore, overexpression of MYH9 in glioblastoma cells reverses the inhibitory effects of CBX7 on migration, proliferation, invasion, and stemness of glioblastoma cells. In summary, CBX7 acts as a tumor suppressor by inhibiting the stem cell-like characteristics of glioblastoma. The CBX7-MYH9-NF-κB signaling axis may serve as a potential therapeutic target for glioblastoma.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: The study was approved by the Institutional Ethics Committee of Wuxi People’s Hospital of Nanjing Medical University at the commencement of the study.
Figures



References
-
- van Wijnen AJ, Bagheri L, Badreldin AA, Larson AN, Dudakovic A, Thaler R, et al. Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development. Bone. 2021;143:115659. - PubMed
-
- Klauke K, Radulovic V, Broekhuis M, Weersing E, Zwart E, Olthof S, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15:353–62. - PubMed
-
- Xiao Q, Wang G, Yin X, Luo Z, Margariti A, Zeng L, et al. Chromobox protein homolog 3 is essential for stem cell differentiation to smooth muscles in vitro and in embryonic arteriogenesis. Arterioscler Thromb Vasc Biol. 2011;31:1842–52. - PubMed
Grants and funding
- BJRC-20/Wuxi Municipal Bureau on Science and Technology (Wuxi Science and Technology Bureau)
- BJ2023013/Wuxi Municipal Bureau on Science and Technology (Wuxi Science and Technology Bureau)
- BK20180169/Wuxi Municipal Bureau on Science and Technology (Wuxi Science and Technology Bureau)
- BJRC-20/Wuxi Municipal Bureau on Science and Technology (Wuxi Science and Technology Bureau)
- BK20180169/Wuxi Municipal Bureau on Science and Technology (Wuxi Science and Technology Bureau)
- BJRC-20/Wuxi Municipal Bureau on Science and Technology (Wuxi Science and Technology Bureau)
- BJ2023013/Wuxi Municipal Bureau on Science and Technology (Wuxi Science and Technology Bureau)
- M2021075/Jiangsu Provincial Commission of Health and Family Planning
- stxqnrc/Jiangsu Provincial Commission of Health and Family Planning
- M2021075/Jiangsu Provincial Commission of Health and Family Planning
- WXKY202304021/Nanjing Medical University (NMU)
- WXKY202404010/Nanjing Medical University (NMU)
- WXKY202404010/Nanjing Medical University (NMU)
LinkOut - more resources
Full Text Sources
Miscellaneous

