Increased ectodysplasin-A2-receptor EDA2R is a ubiquitous hallmark of aging and mediates parainflammatory responses
- PMID: 39988718
- PMCID: PMC11847917
- DOI: 10.1038/s41467-025-56918-3
Increased ectodysplasin-A2-receptor EDA2R is a ubiquitous hallmark of aging and mediates parainflammatory responses
Abstract
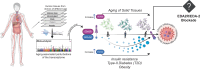
Intensive efforts have been made to identify features that could serve as biomarkers of aging. Yet, drug-based interventions aimed at lessening the detrimental effects of getting older are lacking. This is largely attributable to tissue-specificity, sex-related differences, and to the difficulty of identifying actionable targets, which continues to pose a significant challenge. Here, we implement a bioinformatics approach revealing that aging-associated increase of the transmembrane Ectodysplasin-A2-Receptor is a prominent tissue-independent alteration occurring in humans and other species, and is particularly pronounced in models of accelerated aging. We show that strengthening of the Ectodysplasin-A2-Receptor signalling axis in myogenic precursors and differentiated myotubes suffices to trigger potent parainflammatory responses, mirroring aspects of aging-driven sarcopenia. Intriguingly, obesity, insulin-resistance, and aging-related comorbidities, such as type-2-diabetes, result in heightened levels of the Ectodysplasin-A2 ligand. Our findings suggest that targeting the Ectodysplasin-A2 surface receptor represents a promising pharmacological strategy to mitigate the development of aging-associated phenotypes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Tanikawa, C., Ri, C., Kumar, V., Nakamura, Y. & Matsuda, K. Crosstalk of EDA-A2/XEDAR in the p53 signaling pathway. Mol. Cancer Res.8, 855–863 (2010). - PubMed
-
- Sinha, S. K., Zachariah, S., Quinones, H. I., Shindo, M. & Chaudhary, P. M. Role of TRAF3 and -6 in the activation of the NF-kappa B and JNK pathways by X-linked ectodermal dysplasia receptor. J. Biol. Chem.277, 44953–44961 (2002). - PubMed
-
- Brosh, R. et al. p53-dependent transcriptional regulation of EDA2R and its involvement in chemotherapy-induced hair loss. FEBS Lett.584, 2473–2477 (2010). - PubMed
-
- de Vries, M. et al. Lung tissue gene-expression signature for the ageing lung in COPD. Thorax, 10.1136/thoraxjnl-2017-210074 (2017). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

