Dual-mode action of scalable, high-quality engineered stem cell-derived SIRPα-extracellular vesicles for treating acute liver failure
- PMID: 39988725
- PMCID: PMC11847939
- DOI: 10.1038/s41467-025-57133-w
Dual-mode action of scalable, high-quality engineered stem cell-derived SIRPα-extracellular vesicles for treating acute liver failure
Abstract
Acute liver failure (ALF) is a life-threatening condition caused by rapid hepatocyte death and impaired liver regeneration. Here we show that extracellular vesicles engineered to express Signal Regulatory Protein Alpha (SIRP-EVs), produced via a scalable 3D bioreactor process with high yield and purity, exhibit significant therapeutic potential by targeting damaged cells and promoting tissue repair. SIRP-EVs block CD47, a crucial inhibitory signal on necroptotic cells, to enhance macrophage-mediated clearance of dying hepatocytes. They also deliver regenerative cargo from mesenchymal stem cells, reprogramming macrophages to support liver regeneration. In male animal models, SIRP-EVs significantly reduce liver injury markers and improve survival, demonstrating their dual-function therapeutic efficacy. By integrating the resolution of necroptosis with regenerative macrophage reprogramming, SIRP-EVs represent a promising platform for restoring liver function. These findings support the development of EV-based in vivo macrophage reprogramming therapies for ALF and other inflammation-driven diseases, paving the way for the clinical application of engineered EV therapeutics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: I.-S.K. and G.-H.N. are the co-founders and have stock interest in SHIFTBIO INC. S.K. (Seohyun Kim), Y.K.K., S.K. (Seonghyun Kim), Y.-S.C., I.L., H.J., J.K., Y.C., and G.B.K. are employees of SHIFTBIO INC. T.D.S., J.J., E.Z., and S.L. are employees of RoosterBio, Inc. H.C. are the co-founder and have stock interest in Portrai, Inc. J.P. is an employee of Portrai, Inc. The remaining authors declare no competing interests.
Figures
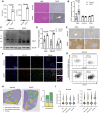
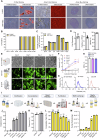

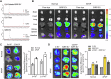

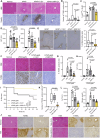


References
-
- Stravitz, R. T. & Kramer, D. J. Management of acute liver failure. Nat. Rev. Gastroenterol. Hepatol.6, 542–553 (2009). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

