Autophagy regulates cellular senescence by mediating the degradation of CDKN1A/p21 and CDKN2A/p16 through SQSTM1/p62-mediated selective autophagy in myxomatous mitral valve degeneration
- PMID: 39988732
- PMCID: PMC12283023
- DOI: 10.1080/15548627.2025.2469315
Autophagy regulates cellular senescence by mediating the degradation of CDKN1A/p21 and CDKN2A/p16 through SQSTM1/p62-mediated selective autophagy in myxomatous mitral valve degeneration
Abstract
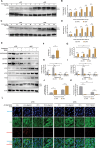
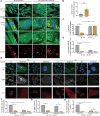
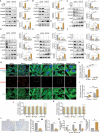
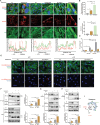
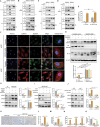
Myxomatous mitral valve degeneration (MMVD) is one of the most important age-dependent degenerative heart valve disorders in both humans and dogs. It is characterized by the aberrant remodeling of extracellular matrix (ECM), regulated by senescent myofibroblasts (aVICs) transitioning from quiescent valve interstitial cells (qVICs), primarily under TGFB1/TGF-β1 control. In the present study, we found senescent aVICs exhibited impaired macroautophagy/autophagy as evidenced by compromised autophagy flux and immature autophagosomes. MTOR-dependent autophagy induced by rapamycin and torin-1 attenuated cell senescence and decreased the expression of cyclin-dependent kinase inhibitors (CDKIs) CDKN2A/p16INK4A and CDKN1A/p21CIP1. Furthermore, induction of autophagy in aVICs by ATG (autophagy related) gene overexpression restored autophagy flux, with a concomitant reduction in CDKN1A and CDKN2A expression and senescence-associated secretory phenotype (SASP). Conversely, autophagy deficiency induced CDKN1A and CDKN2A accumulation and SASP, whereas ATG re-expression alleviated senescent phenotypic transformation. Notably, CDKN1A and CDKN2A localized to autophagosomes and lysosomes following MTOR antagonism or MG132 treatment. SQSTM1/p62 was identified as the autophagy receptor to selectively sequester CDKN1A and CDKN2A cargoes for autophagic degradation. Our findings are the first demonstration that CDKN1A and CDKN2A are degraded through SQSTM1-mediated selective autophagy, independent of the ubiquitin-proteasome pathway. These data will inform development of therapeutic strategies for the treatment of canine and human MMVD, and for the treatment of Alzheimer disease, Parkinson disease and other age-related degenerative disorders.Abbreviations: ACTA2/α-SMA: actin alpha 2, smooth muscle; AKT: AKT serine/threonine kinase; aVICs: activated valve interstitial cells; ATG: autophagy related; baf-A1: bafilomycin A1; BrdU, bromodeoxyuridine; BSA: bovine serum albumin; CDKIs, cyclin-dependent kinase inhibitors; CDKN1A/p21: cyclin dependent kinase inhibitor 1A; CDKN2A/p16: cyclin dependent kinase inhibitor 2A; co-IP: co-immunoprecipitation; DMSO: dimethylsulfoxide; ECM, extracellular matrix; EIF4EBP1: eukaryotic translation initiation factor 4E binding protein 1; eGFP: green fluorescent protein; ELISA: enzyme-linked immunosorbent assay; HEK-293T, human embryonic kidney 293T; HRP: horseradish peroxidase; KO: knockout; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; LIR: MAP1LC3/LC3-interacting region; MFS: Marfan syndrome; MKI67/Ki-67: marker of proliferation Ki-67; MMVD: myxomatous mitral valve degeneration; MTOR: mechanistic target of rapamycin kinase; MTORC: MTOR complex; OE: overexpression; PBST, phosphate-buffered saline with 0.1% Tween-20; PCNA: proliferating cell nuclear antigen; PIK3CA/PI3K: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PLA: proximity ligation assays; PSMA1: proteasome 20S subunit alpha 1; PSMB5: proteasome 20S subunit beta 5; qVICs: quiescent valve interstitial cells; qRT-PCR: quantitative real-time PCR; SA-GLB1/β-gal: SA-senescence-associated GLB1/β-galactosidase; ROS: reactive oxygen species; SASP: senescence-associated secretory phenotype; RPS6KB1/p70 S6K: ribosomal protein S6 kinase B1; SMAD: SMAD family member; SQSTM1/p62: sequestosome 1; STEM: scanning transmission electron microscopy; TGFB: transforming growth factor beta; TGFBR: transforming growth factor beta receptor; TP53/p53: tumor protein p53; UPS: ubiquitin-proteasome system; WT, wild-type.
Keywords: Autophagic degradation; CDKI; MMVD; SASP; TGFB; ubiquitin-proteasome pathway.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
