Genomic surveillance and evolution of Getah virus
- PMID: 39989716
- PMCID: PMC11844246
- DOI: 10.1093/ve/veaf007
Genomic surveillance and evolution of Getah virus
Abstract
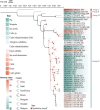
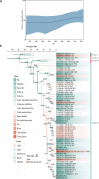
Getah virus (GETV), a member of the Alphaviruses, has spread widely and is expanding its host range worldwide, posing a serious threat to public health safety and the farming industry. However, genetic monitoring of GETV is inadequate, and its evolution and transmission remain unclear. This study employed reverse transcription-polymerase chain reaction to screen pig tissue samples for the presence of GETV. Subsequent steps included DNA sequencing, phylogenetic analysis, and selection pressure assessments to elucidate the evolutionary history and transmission patterns of the virus. A total of 1382 samples were examined, with a positivity rate of 4.12% (95% confidence interval: 3.07%-5.17%) from 2022 to 2023. Subsequently, seven GETV strains were isolated and identified. A phylogenetic tree was constructed, which showed that all seven strains belonged to Group III. Phylodynamic analysis revealed that GETV evolved rapidly. Additionally, eight amino acid sites within the GETV E2 protein were identified as being under positive selection. These data provide insight into the epidemiology and evolution of GETV.
Keywords: Getah virus; alphavirus; epidemiology; evolution; phylogenetic dynamics.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
This manuscript has been reviewed and approved by all authors. There are no commercial or financial relationships that would lead to a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

