This is a preprint.
Air sampling accurately captures circulating zoonotic viral diversity emerging from poultry live-animal markets
- PMID: 39989955
- PMCID: PMC11844658
- DOI: 10.21203/rs.3.rs-5682962/v1
Air sampling accurately captures circulating zoonotic viral diversity emerging from poultry live-animal markets
Abstract
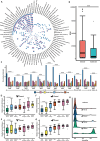
Environmental surveillance has emerged as a pivotal strategy for early detection of pathogens that pose threats to humans (1) but has not been utilized for zoonotic agents. In Asia, live-bird markets (LBMs) are key human-animal interfaces for zoonotic virus transmission (2). Traditional sampling strategies are time-consuming, expensive, threaten animal welfare and have significant occupational biosafety risks. In this study, we assessed the performance of metagenomics on environmental samples (ES) compared to traditional poultry swabs for detecting avian viral pathogens in LBMs in Cambodia. ES, including air, cage swabs, and carcass wash water, were collected alongside throat and cloacal swabs from domestic chickens and ducks across twelve sampling visits in two LBMs over a 15-month period. Viral nucleic acids were extracted and sequenced using a capture probe-based metagenomics approach. Our results show that metagenomics on ES outperformed traditional poultry samples in detecting the highly pathogenic Influenza A/H5N1, including circulating clades 2.3.4.4b and 2.3.2.1c, which were found in the environment but missed by poultry swabs on multiple occasions. Environmental metagenomics was also highly sensitive in the detection of over 40 other viruses from key pathogen families such as Astroviridae, Coronaviridae, Picornaviridae, and Retroviridae. Viral contigs from ES showed high similarity to those from poultry swabs further highlighting the accuracy of this approach. Our findings highlight metagenomics on ES can precisely and effectively replicate metagenomic results from traditional surveillance samples, offering broader coverage and enhanced detection of avian pathogens. This robust approach could be pivotal for mitigating zoonotic spillover, controlling pathogen transmission at LBMs, and enhancing pandemic preparedness strategies.
Conflict of interest statement
Competing interests The authors declare no competing interests. Declarations Ethics statement All sample collections conducted for this study adhered to ethical guidelines and were carried out in collaboration with the National Animal Health and Production Institute. The collections were conducted under protocols approved by the National Ethics Committee for Health Research (NECHR) under the Ministry of Health, Cambodia, with approval numbers NECHR013, NECHR143, NECHR149, and NECHR320 approved in 2021 and 2022. Cambodia does not have a specific animal ethics review board; however, further informal approvals for animal sample collections are given by the General Directorate of Animal Health and Production under the Ministry of Agriculture, Forestry and Fisheries.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

