This is a preprint.
Cross-Ancestry Comparison of Aptamer and Antibody Proteomics Measures
- PMID: 39989965
- PMCID: PMC11844639
- DOI: 10.21203/rs.3.rs-5968391/v1
Cross-Ancestry Comparison of Aptamer and Antibody Proteomics Measures
Abstract
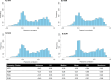
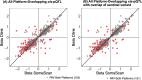
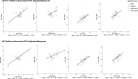
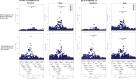
Measures from affinity-proteomics platforms often correlate poorly, challenging interpretation of protein associations with genetic variants (pQTL) and phenotypes. Here, we examined 2,157 proteins measured on both SomaScan 7k and Olink Explore 3072 across 1,930 participants with genetic similarity to European, African, East Asian, and Admixed American ancestry references. Inter-platform correlation coefficients for these 2,157 proteins followed a bimodal distribution (median r=0.30). Protein measures from each platform were associated with genetic variants (pQTLs), and one-third of the pQTL signals were driven by protein-altering variants (PAVs). We highlight 80 proteins that correlate differently across ancestry groups likely due to differing PAV frequencies by ancestry. Furthermore, adjustment for PAVs with opposite directions of effect by platform improved inter-platform protein measure correlation and resulted in more concordant genetic and phenotypic associations. Hence, PAVs need to be accounted for across ancestries to facilitate platform-concordant and accurate protein measurement.
Conflict of interest statement
Conflicts of interest LMR is a consultant for the TOPMed Administrative Coordinating Center (through Westat). DG is the Chief Executive Officer of Sequoia Genetics, a private company that works with investors, pharma, biotech, and academia by performing research that leverages genetic data to help inform drug discovery and development. DG has interests in several biotech companies.
Figures









References
-
- Ferkingstad E. et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 53, 1712–1721 (2021). - PubMed
-
- Said S. et al. Ancestry diversity in the genetic determinants of the human plasma proteome and associated new drug targets. medRxiv (2023) doi:10.1101/2023.11.13.23298365. - DOI
Publication types
Grants and funding
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R01 HL071259/HL/NHLBI NIH HHS/United States
- T32 ES007018/ES/NIEHS NIH HHS/United States
- 75N92020D00001/HL/NHLBI NIH HHS/United States
- R01 HL071250/HL/NHLBI NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- OT3 HL142481/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 HL159081/HL/NHLBI NIH HHS/United States
- R01 HL071205/HL/NHLBI NIH HHS/United States
- R01 HL071258/HL/NHLBI NIH HHS/United States
- N01 HC095160/HL/NHLBI NIH HHS/United States
- OT3 HL142478/HL/NHLBI NIH HHS/United States
- UL1 RR033176/RR/NCRR NIH HHS/United States
- 75N92020D00002/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- T32 GM135128/GM/NIGMS NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL071251/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- R01 DK072193/DK/NIDDK NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- OT3 HL147154/HL/NHLBI NIH HHS/United States
- 75N92020D00003/HL/NHLBI NIH HHS/United States
- R01 HL146860/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 HL071051/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- U01 AG082042/AG/NIA NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- 75N92020D00004/HL/NHLBI NIH HHS/United States
- OT3 HL142480/HL/NHLBI NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- 75N92020D00007/HL/NHLBI NIH HHS/United States
- HHSN268201500014C/HL/NHLBI NIH HHS/United States
- R01 HL133870/HL/NHLBI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- OT3 HL142479/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- R01 HL151855/HL/NHLBI NIH HHS/United States
- UM1 DK078616/DK/NIDDK NIH HHS/United States
- 75N92020D00006/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

