Calnexin promotes glioblastoma progression by inducing protective mitophagy through the MEK/ERK/BNIP3 pathway
- PMID: 39990231
- PMCID: PMC11840740
- DOI: 10.7150/thno.105591
Calnexin promotes glioblastoma progression by inducing protective mitophagy through the MEK/ERK/BNIP3 pathway
Abstract
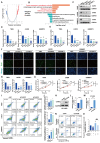
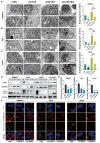
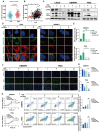
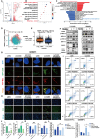
Rationale: Glioblastoma multiforme (GBM), one of the most malignant tumors of the central nervous system, has a poor prognosis, mainly because of its high recurrence caused by the rapid development of drug resistance to postoperative chemotherapy. Although macroautophagy/autophagy is believed to be a fundamental factor in tumor survival during chemotherapy, there is still a lack of autophagy biomarkers for predicting patient prognosis and chemotherapeutic efficacy in clinical practice. Methods: We combined transcriptomic and single-cell sequencing data to identify differentially expressed autophagy-related genes in gliomas. Overexpression of calnexin (CANX), a key gene related to protein folding, and its secretion in the endoplasmic reticulum (ER) was identified, suggesting poor prognosis in GBM patients. The autophagy flow related to CANX was detected by transmission electron microscopy (TEM), Western blotting, and immunofluorescence. Flow cytometry, cell proliferation, activity assays, and the GBM intracranial xenograft mouse model were employed to validate CANX's role in GBM progression. Results: CANX knockdown inhibited proliferation and autophagosome formation in GBM cells. On the other hand, CANX overexpression increased mitogen-activated protein kinase (MAPK) activity, leading to the accumulation of BNIP3 (CL2/adenovirus E1B 19 kDa interacting protein 3, a critical factor regulating mitophagy) and protective mitophagy. Notably, when combined with temozolomide (TMZ), CANX knockdown extended the lifespan of GBM-bearing mice. Additionally, our studies revealed that the classic calcium inhibitor nimodipine (ND) decreased CANX expression and thus enhanced the sensitivity to TMZ. Conclusions: Our findings indicate that CANX functions as an oncogene in GBM. We also characterize the CANX/MEK/ERK/BNIP3 mitophagy pathway, provide new insights into the molecular mechanism of GBM drug resistance, and identify a therapeutic target.
Keywords: calnexin; glioblastoma; mitophagy; nimodipine; temozolomide.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. - PubMed
-
- Rizzo AE, Yu JS. Radiation therapy for glioma stem cells. Adv Exp Med Biol. 2015;853:85–110. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

