This is a preprint.
Mutational scanning of TnpB reveals latent activity for genome editing
- PMID: 39990302
- PMCID: PMC11844463
- DOI: 10.1101/2025.02.11.637750
Mutational scanning of TnpB reveals latent activity for genome editing
Abstract
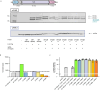
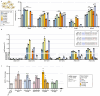
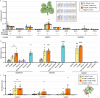
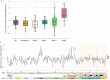
TnpB is a diverse family of RNA-guided endonucleases associated with prokaryotic transposons. Due to their small size and putative evolutionary relationship to CRISPR-Cas12, TnpB enzymes hold significant potential for genome editing. However, most TnpBs lack robust gene editing activity, and unbiased profiling of mutational effects on editing activity has not been explored. Here, we mapped comprehensive sequence-function landscapes of a TnpB ribonucleoprotein and discovered many activating mutations in both the protein and RNA. One- and two-position RNA mutants outperform existing variants, highlighting the utility of systematic RNA scaffold mutagenesis. Leveraging the protein's mutational landscape, we identified enhanced TnpB variants from a combinatorial library of activating mutations. These variants enhanced editing in human cells, N. benthamiana, pepper, and rice, with up to a fifty-fold increase compared to wild-type TnpB. These findings highlight previously unknown elements critical for regulating TnpB endonuclease activity and reveal surprising latent activity accessible through mutation.
Figures










References
Methods References
-
- Stuckey S. & Storici F. Gene knockouts, in vivo site-directed mutagenesis and other modifications using the delitto perfetto system in Saccharomyces cerevisiae. Methods Enzymol. 533, 103–131 (2013). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
