This is a preprint.
Identification of a multi-omics factor predictive of long COVID in the IMPACC study
- PMID: 39990442
- PMCID: PMC11844572
- DOI: 10.1101/2025.02.12.637926
Identification of a multi-omics factor predictive of long COVID in the IMPACC study
Update in
-
A multiomics recovery factor predicts long COVID in the IMPACC study.J Clin Invest. 2025 Sep 9;135(21):e193698. doi: 10.1172/JCI193698. eCollection 2025 Nov 3. J Clin Invest. 2025. PMID: 40924481 Free PMC article. Clinical Trial.
Abstract
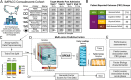
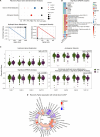
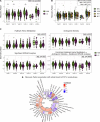
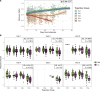
Following SARS-CoV-2 infection, ~10-35% of COVID-19 patients experience long COVID (LC), in which often debilitating symptoms persist for at least three months. Elucidating the biologic underpinnings of LC could identify therapeutic opportunities. We utilized machine learning methods on biologic analytes and patient reported outcome surveys provided over 12 months after hospital discharge from >500 hospitalized COVID-19 patients in the IMPACC cohort to identify a multi-omics "recovery factor". IMPACC participants who experienced LC had lower recovery factor scores compared to participants without LC. Biologic characterization revealed increased levels of plasma proteins associated with inflammation, elevated transcriptional signatures of heme metabolism, and decreased androgenic steroids in LC patients. The recovery factor was also associated with altered circulating immune cell frequencies. Notably, recovery factor scores were predictive of LC occurrence in patients as early as hospital admission, irrespective of acute disease severity. Thus, the recovery factor identifies patients at risk of LC early after SARS-CoV-2 infection and reveals LC biomarkers and potential treatment targets.
Keywords: COVID-19; Heme metabolism; Long COVID; Machine Learning; PASC; SARS-CoV-2; androgenic steroids; inflammation; multi-omics; patient reported outcomes.
Conflict of interest statement
The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays, NDV-based SARS-CoV-2 vaccines influenza virus vaccines and influenza virus therapeutics which lists Florian Krammer as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2 and another company, Castlevax, to develop SARS-CoV-2 vaccines. Florian Krammer is co-founder and scientific advisory board member of Castlevax. Florian Krammer has consulted for Merck, Curevac, Seqirus, GSK and Pfizer and is currently consulting for 3rd Rock Ventures, Sanofi, Gritstone and Avimex. The Krammer laboratory is also collaborating with Dynavax on influenza vaccine development and with VIR on influenza virus therapeutics development. Viviana Simon is a co-inventor on a patent filed relating to SARS-CoV-2 serological assays (the “Serology Assays”). Ofer Levy is a named inventor on patents held by Boston Children’s Hospital relating to vaccine adjuvants and human in vitro platforms that model vaccine action. His laboratory has received research support from GlaxoSmithKline (GSK) and Pfizer, and he is a co-founder of and advisor to Ovax, Inc that develops opioid vaccines. Charles Cairns serves as a consultant to bioMerieux and is funded by a grant from Bill & Melinda Gates Foundation. James A Overton is a consultant at Knocean Inc. Jessica Lasky-Su serves as a scientific advisor of Precion Inc. Scott R. Hutton, Greg Michelloti and Kari Wong are employees of Metabolon Inc. Vicki Seyfer- Margolis is a current employee of MyOwnMed. Nadine Rouphael reports grants or contracts with Merck, Sanofi, Pfizer, Vaccine Company and Immorna, and has participated on data safety monitoring boards for Moderna, Sanofi, Seqirus, Pfizer, EMMES, ICON, BARDA, and CyanVan, Imunon Micron. N.R. has also received support for meetings/travel from Sanofi and Moderna and honoraria from Virology Education and Krog consulting. Chris Cotsapas is a current employee of Vesalius Therapeutics. Adeeb Rahman is a current employee of Immunai Inc. Steven Kleinstein is a consultant related to ImmPort data repository for Peraton. Nathan Grabaugh is a consultant for Tempus Labs and the National Basketball Association. Akiko Iwasaki is a consultant for 4BIO, Blue Willow Biologics, Revelar Biotherapeutics, RIGImmune, Xanadu Bio, Paratus Sciences. Monika Kraft receives research funds paid to her institution from NIH, ALA; Sanofi, Astra-Zeneca for work in asthma, serves as a consultant for Astra-Zeneca, Sanofi, Chiesi, GSK for severe asthma; is a co-founder and CMO for RaeSedo, Inc, a company created to develop peptidomimetics for treatment of inflammatory lung disease. Esther Melamed received research funding from Babson Diagnostics and honorarium from Multiple Sclerosis Association of America and has served on the advisory boards of Genentech, Horizon, Teva, and Viela Bio. Carolyn Calfee receives research funding from NIH, FDA, DOD, Roche-Genentech and Quantum Leap Healthcare Collaborative as well as consulting services for Janssen, Vasomune, Gen1e Life Sciences, NGMBio, and Cellenkos. Wade Schulz was an investigator for a research agreement, through Yale University, from the Shenzhen Center for Health Information for work to advance intelligent disease prevention and health promotion; collaborates with the National Center for Cardiovascular Diseases in Beijing; is a technical consultant to Hugo Health, a personal health information platform; cofounder of Refactor Health, an AI-augmented data management platform for health care; and has received grants from Merck and Regeneron Pharmaceutical for research related to COVID-19. Grace A McComsey received research grants from Rehdhill, Cognivue, Pfizer, and Genentech, and served as a research consultant for Gilead, Merck, Viiv/GSK, and Janssen. Linda N. Geng received research funding paid to her institution from Pfizer, Inc. Catherine Hough receives support through her institution from the NIH and CDC. David Hafler has received research funding from BristolMyers Squibb, Novartis, Sanofi, and Genentech. He has been a consultant for Bayer Pharmaceuticals, Repertoire Inc, Bristol Myers Squibb, Compass Therapeutics, EMD Serono, Genentech, Novartis Pharmaceuticals, and Sanofi Genzyme.
Figures


References
-
- Groff D., Sun A., Ssentongo A.E., Ba D.M., Parsons N., Poudel G.R., Lekoubou A., Oh J.S., Ericson J.E., Ssentongo P., et al. (2021). Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection: A Systematic Review. JAMA Netw Open 4, e2128568. 10.1001/jamanetworkopen.2021.28568. - DOI - PMC - PubMed
-
- Melamed E., Rydberg L., Ambrose A.F., Bhavaraju-Sanka R., Fine J.S., Fleming T.K., Herman E., Phipps Johnson J.L., Kucera J.R., Longo M., et al. (2023). Multidisciplinary collaborative consensus guidance statement on the assessment and treatment of neurologic sequelae in patients with post-acute sequelae of SARS-CoV-2 infection (PASC). PM R 15, 640–662. 10.1002/pmrj.12976. - DOI - PubMed
Publication types
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U01 AI167892/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- T32 DA018926/DA/NIDA NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- R01 AI122220/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- UM1 TR004528/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
