This is a preprint.
Cohesin drives chromatin scanning during the RAD51-mediated homology search
- PMID: 39990468
- PMCID: PMC11844420
- DOI: 10.1101/2025.02.10.637451
Cohesin drives chromatin scanning during the RAD51-mediated homology search
Update in
-
Cohesin drives chromatin scanning during the RAD51-mediated homology search.Science. 2025 Dec 4;390(6777):eadw1928. doi: 10.1126/science.adw1928. Epub 2025 Dec 4. Science. 2025. PMID: 41343630
Abstract
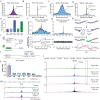
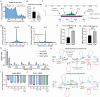
Cohesin folds genomes into chromatin loops, whose roles are under debate. We report that double strand breaks (DSB) induce de novo formation of chromatin loops, with the break positioned at the loop base. These loops form only in S/G2 phases and occur during repair via homologous recombination (HR), concomitant with DNA end resection and RAD51 assembly. RAD51 showed two-tiered accumulation around DSBs, with a broad (~Mb) domain arising from the homology search. This domain is regulated by cohesin unloader, is constrained by TAD boundaries, and it overlaps with chromatin regions reeled through the break-anchored loop, suggesting that loop extrusion regulates the homology search. Indeed, depletion of NIPBL results in reduced HR, and this effect is more pronounced when the HR donor is far (~100 kb) from the break. Our data indicates that loop-extruding cohesin promotes the mammalian homology search by facilitating break-chromatin interactions within the damaged TAD.
Conflict of interest statement
Competing interests: Authors declare that they have no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
