This is a preprint.
Machine learning models predict long COVID outcomes based on baseline clinical and immunologic factors
- PMID: 39990570
- PMCID: PMC11844586
- DOI: 10.1101/2025.02.12.25322164
Machine learning models predict long COVID outcomes based on baseline clinical and immunologic factors
Abstract
The post-acute sequelae of SARS-CoV-2 (PASC), also known as long COVID, remain a significant health issue that is incompletely understood. Predicting which acutely infected individuals will go on to develop long COVID is challenging due to the lack of established biomarkers, clear disease mechanisms, or well-defined sub-phenotypes. Machine learning (ML) models offer the potential to address this by leveraging clinical data to enhance diagnostic precision. We utilized clinical data, including antibody titers and viral load measurements collected at the time of hospital admission, to predict the likelihood of acute COVID-19 progressing to long COVID. Our machine learning models achieved median AUROC values ranging from 0.64 to 0.66 and AUPRC values between 0.51 and 0.54, demonstrating their predictive capabilities. Feature importance analysis revealed that low antibody titers and high viral loads at hospital admission were the strongest predictors of long COVID outcomes. Comorbidities, including chronic respiratory, cardiac, and neurologic diseases, as well as female sex, were also identified as significant risk factors for long COVID. Our findings suggest that ML models have the potential to identify patients at risk for developing long COVID based on baseline clinical characteristics. These models can help guide early interventions, improving patient outcomes and mitigating the long-term public health impacts of SARS-CoV-2.
Keywords: COVID-19; Long COVID; PASC; Patient Reported Outcomes; SARS-CoV-2; modeling.
Conflict of interest statement
IMPACC Network Competing Interests The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list Florian Krammer as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. Florian Krammer has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Seqirus, 3rd Rock Ventures, Merck and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2. Viviana Simon is a co-inventor on a patent filed relating to SARS-CoV-2 serological assays (the “Serology Assays”). Ofer Levy is a named inventor on patents held by Boston Children’s Hospital relating to vaccine adjuvants and human in vitro platforms that model vaccine action. His laboratory has received research support from GlaxoSmithKline (GSK). Charles Cairns serves as a consultant to bioMerieux and is funded for a grant from Bill & Melinda Gates Foundation. James A Overton is a consultant at Knocean Inc. Jessica Lasky-Su serves as a scientific advisor of Precion Inc. Scott R. Hutton, Greg Michelloti and Kari Wong are employees of Metabolon Inc. Vicki Seyfer-Margolis is a current employee of MyOwnMed. Nadine Rouphael reports contracts with Lilly,Immorna, Vaccine Company and Sanofi for COVID-19 clinical trials and serves as a consultant for ICON, EMMES, Imunon, CyanVac for consulting on safety for COVID19 clinical trials. Adeeb Rahman is a current employee of Immunai Inc. Steven Kleinstein is a consultant related to ImmPort data repository for Peraton. Nathan Grabaugh is a consultant for Tempus Labs and the National Basketball Association. Akiko Iwasaki is a consultant for 4BIO, Blue Willow Biologics, Revelar Biotherapeutics, RIGImmune, Xanadu Bio, Paratus Sciences. Monika Kraft receives research funds paid to her institution from NIH, ALA; Sanofi, Astra-Zeneca for work in asthma, serves as a consultant for Astra-Zeneca, Sanofi, Chiesi, GSK for severe asthma; is a co-founder and CMO for RaeSedo, Inc, a company created to develop peptidomimetics for treatment of inflammatory lung disease. Esther Melamed received research funding from Babson Diagnostics, honorarium from Multiple Sclerosis Association of America and has served on advisory boards of Genentech, Horizon, Teva and Viela Bio. Carolyn Calfee receives research funding from NIH, FDA, DOD, Roche-Genentech and Quantum Leap Healthcare Collaborative as well as consulting services for Janssen, Vasomune, Gen1e Life Sciences, NGMBio, and Cellenkos. Wade Schulz was an investigator for a research agreement, through Yale University, from the Shenzhen Center for Health Information for work to advance intelligent disease prevention and health promotion; collaborates with the National Center for Cardiovascular Diseases in Beijing; is a technical consultant to Hugo Health, a personal health information platform; cofounder of Refactor Health, an AI-augmented data management platform for health care; and has received grants from Merck and Regeneron Pharmaceutical for research related to COVID-19. Grace A McComsey received research grants from Rehdhill, Cognivue, Pfizer, and Genentech, and served as a research consultant for Gilead, Merck, Viiv/GSK, and Jenssen. Linda N. Geng received research funding paid to her institution from Pfizer, Inc.
Figures
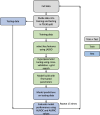


References
-
- National Center for Health Statistics. U.S. Census Bureau HPS, 2022–. Long COVID; n.d.
-
- Office for National Statistics. Prevalence of ongoing symptoms following coronavirus (COVID-19) infection in the UK; n.d.
Publication types
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- S10 OD026799/OD/NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- R01 AI132774/AI/NIAID NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
