A Reverse Transcription Nucleic-Acid-Based Barcoding System for In Vivo Measurement of Lipid Nanoparticle mRNA Delivery
- PMID: 39990951
- PMCID: PMC11843327
- DOI: 10.1021/acsbiomedchemau.4c00081
A Reverse Transcription Nucleic-Acid-Based Barcoding System for In Vivo Measurement of Lipid Nanoparticle mRNA Delivery
Abstract
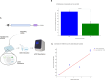
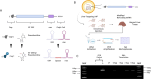
Lipid nanoparticles (LNPs) are the most extensively validated clinical delivery vehicles for mRNA therapeutics, exemplified by their widespread use in the mRNA COVID-19 vaccines. The pace of lipid nanoparticle (LNP) development for mRNA therapeutics is restricted by the limitations of existing methods for large-scale LNP screening. To address this challenge, we developed Quantitative Analysis of Reverse Transcribed Barcodes (QuART), a novel nucleic-acid-based system for measuring LNP functional delivery in vivo. QuART uses a bacterial retron reverse transcription system to couple functional mRNA delivery into the cytoplasm with a cDNA barcode readout. Our results demonstrate that QuART can be used to identify functional mRNA delivery both in vitro in cell culture and in vivo in mice. Multiplexing of QuART could enable high-throughput screening of LNP formulations, facilitating the rapid discovery of promising LNP candidates for mRNA therapeutics.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
LinkOut - more resources
Full Text Sources
