Advanced delivery systems for gene editing: A comprehensive review from the GenE-HumDi COST Action Working Group
- PMID: 39991472
- PMCID: PMC11847086
- DOI: 10.1016/j.omtn.2025.102457
Advanced delivery systems for gene editing: A comprehensive review from the GenE-HumDi COST Action Working Group
Abstract
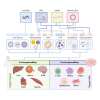
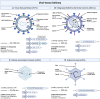
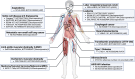
In the past decade, precise targeting through genome editing has emerged as a promising alternative to traditional therapeutic approaches. Genome editing can be performed using various platforms, where programmable DNA nucleases create permanent genetic changes at specific genomic locations due to their ability to recognize precise DNA sequences. Clinical application of this technology requires the delivery of the editing reagents to transplantable cells ex vivo or to tissues and organs for in vivo approaches, often representing a barrier to achieving the desired editing efficiency and safety. In this review, authored by members of the GenE-HumDi European Cooperation in Science and Technology (COST) Action, we described the plethora of delivery systems available for genome-editing components, including viral and non-viral systems, highlighting their advantages, limitations, and potential application in a clinical setting.
Keywords: COST; European Cooperation in Science and Technology; GenE-HumDi; MT: Delivery Strategies; base editors; delivery systems; genome editing; regulatory guidelines.
© 2025 The Author(s).
Conflict of interest statement
The authors disclose being members of the GenE-HumDi COST Action CA21113. A.C. has licensed medicinal products and receives patents and royalties from Danaus Pharmaceuticals. A.C. is inventor on a patent for MEGA gene editing off-target detection (WO/2023/079285). P.R. has licensed medicinal products and receives funding and equity from Rocket Pharmaceuticals, Inc.; patents and royalties, and research and consulting funding.
Figures


References
-
- Wang J.Y., Doudna J.A. CRISPR technology: A decade of genome editing is only the beginning. Science. 2023;379 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

