Germline Pathogenic DROSHA Variants Are Linked to Pineoblastoma and Wilms Tumor Predisposition
- PMID: 39992227
- PMCID: PMC11995001
- DOI: 10.1158/1078-0432.CCR-24-2785
Germline Pathogenic DROSHA Variants Are Linked to Pineoblastoma and Wilms Tumor Predisposition
Abstract
Purpose: DROSHA, DGCR8, and DICER1 regulate miRNA biogenesis and are commonly mutated in cancer. Although DGCR8 and DICER1 germline pathogenic variants (GPV) cause autosomal dominant tumor predisposition, no association between DROSHA GPVs and clinical phenotypes has been reported.
Experimental design: After obtaining informed consent, sequencing was performed on germline and tumor samples from all patients. The occurrence of germline DROSHA GPVs was investigated in large pediatric and adult cancer datasets. The population prevalence of DROSHA GPVs was investigated in the UK Biobank and Geisinger DiscovEHR cohorts.
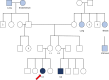
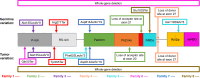
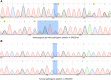
Results: We describe nine children from eight families with heterozygous DROSHA GPVs and a diagnosis of pineoblastoma (n = 8) or Wilms tumor (n = 1). A somatic second hit in DROSHA was detected in all eight tumors analyzed. All pineoblastoma tumors analyzed were classified as miRNA processing-altered 1 subtype. We estimate the population prevalence of germline DROSHA loss-of-function variants to be 1:3,875 to 1:4,843 but find no evidence for increased adult cancer risk.
Conclusions: This is the first report of DROSHA-related tumor predisposition. As pineoblastoma and Wilms tumor are also associated with DICER1 GPVs, our results suggest that the tissues of origin for these tumors are uniquely tolerant of general miRNA loss. The miRNA processing-altered 1 pineoblastoma subtype is associated with older age of diagnosis and better outcomes than other subtypes, suggesting DROSHA GPV status may have important clinical and prognostic significance. We suggest that genetic testing for DROSHA GPVs be considered for patients with pineoblastoma, Wilms tumor, or other DICER1-/DGCR8-related conditions and propose surveillance recommendations through research studies for individuals with DROSHA GPVs.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
S.A. Jackson reports current employment with Natera and ownership of Natera stocks as well as past employment with Invitae and past ownership of Invitae stocks. D.J. Carey reports grants from the NIH during the conduct of the study. S.E. Plon reports she is a member of the Scientific Advisory Panel of Baylor Genetics. K.A.P. Schultz reports other support from the Pine Tree Apple Classic Fund and Children’s Minnesota Foundation during the conduct of the study. No disclosures were reported by the other authors.
Figures


References
-
- Han J, Lee Y, Yeom K-H, Nam J-W, Heo I, Rhee J-K, et al. . Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006;125:887–901. - PubMed
-
- Pelletier D, Rivera B, Fabian MR, Foulkes WD. miRNA biogenesis and inherited disorders: clinico-molecular insights. Trends Genet 2023;39:401–14. - PubMed
-
- Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, et al. . Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 2012;366:234–42. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

