CTCF regulates global chromatin accessibility and transcription during rod photoreceptor development
- PMID: 39993185
- PMCID: PMC11892594
- DOI: 10.1073/pnas.2416384122
CTCF regulates global chromatin accessibility and transcription during rod photoreceptor development
Abstract
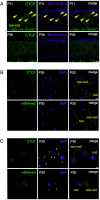
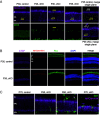
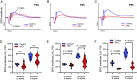
Chromatin architecture facilitates accurate transcription at a number of loci, but it remains unclear how much chromatin architecture is involved in global transcriptional regulation. Previous work has shown that rapid depletion of the architectural protein CTCF in cell culture alters global chromatin organization but results in surprisingly limited gene expression changes. This discrepancy has also been observed when other architectural proteins are depleted, and one possible explanation is that full transcriptional changes are masked by cellular heterogeneity. We tested this idea by performing multiomics analyses with sorted juvenile postmitotic mouse rods, which undergo synchronized development, and we identified CTCF-dependent regulation of global chromatin accessibility and gene expression. CTCF depletion leads to dysregulation of ~20% of the entire transcriptome (>3,000 genes) and ~41% of genome accessibility (>27,000 sites) before any prominent cellular or physiological phenotypes arise. Importantly, these changes are highly enriched for CTCF occupancy at euchromatin, suggesting direct CTCF binding and transcriptional regulation at these active loci. CTCF mainly promotes chromatin accessibility and frequently inhibits expression of these direct binding targets, which are enriched for binding motifs of transcription repressors. These findings provide different and sometimes opposite conclusions from previous studies, emphasizing the need to consider cellular heterogeneity and cell-type specificity when performing multiomics analyses. CTCF knockout rods undergo complete degeneration by adulthood, indicating an essential role for their viability. We conclude that the architectural protein CTCF binds chromatin and regulates global chromatin accessibility and transcription during rod development.
Keywords: CTCF; chromatin architecture; retinal development; rod; transcriptome.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Update of
-
CTCF regulates global chromatin accessibility and transcription during rod photoreceptor development.bioRxiv [Preprint]. 2024 May 31:2024.05.27.596084. doi: 10.1101/2024.05.27.596084. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 Mar 04;122(9):e2416384122. doi: 10.1073/pnas.2416384122. PMID: 38853900 Free PMC article. Updated. Preprint.
References
-
- Hirayama T., et al. , CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2, 345–357 (2012). - PubMed
-
- Sams D. S., et al. , Neuronal CTCF is necessary for basal and experience-dependent gene regulation, memory formation, and genomic structure of BDNF and Arc. Cell Rep. 17, 2418–2430 (2016). - PubMed
MeSH terms
Substances
Grants and funding
- R00 HD097308/HD/NICHD NIH HHS/United States
- startup/University at Buffalo (UB)
- DK015602/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- 4R00HD097308-02/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- K99HD097308-01A1/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

