Insights from a methylome-wide association study of antidepressant exposure
- PMID: 39994233
- PMCID: PMC11850842
- DOI: 10.1038/s41467-024-55356-x
Insights from a methylome-wide association study of antidepressant exposure
Abstract
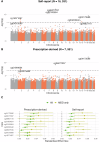
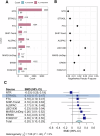
This study tests the association of whole-blood DNA methylation and antidepressant exposure in 16,531 individuals from Generation Scotland (GS), using self-report and prescription-derived measures. We identify 8 associations and a high concordance of results between self-report and prescription-derived measures. Sex-stratified analyses observe nominally significant increased effect estimates in females for four CpGs. There is observed enrichment for genes expressed in the Amygdala and annotated to synaptic vesicle membrane ontology. Two CpGs (cg15071067; DGUOK-AS1 and cg26277237; KANK1) show correlation between DNA methylation with the time in treatment. There is a significant overlap in the top 1% of CpGs with another independent methylome-wide association study of antidepressant exposure. Finally, a methylation profile score trained on this sample shows a significant association with antidepressant exposure in a meta-analysis of eight independent external datasets. In this large investigation of antidepressant exposure and DNA methylation, we demonstrate robust associations which warrant further investigation to inform on the design of more effective and tolerated treatments for depression.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.E.M. is an advisor to the Epigenetic Clock Development Foundation and Optima Partners. D.L.M. was employed by Optima Partners Ltd in a part-time capacity. H.J.G. has received travel grants and speaker honoraria from Fresenius Medical Care, Neuraxpharm, Servier, Indorsia and Janssen Cilag, as well as research funding from Fresenius Medical Care. H.J.G. had personal contracts approved by the university administration for speaker honoraria and one IIT with Fresenius Medical Care. T.K. received unrestricted educational grants from Servier, Janssen, Recordati, Aristo, Otsuka, neuraxpharm. B.W.J.H.P. has received research funding (not related to the current paper) from Boehringer Ingelheim and Jansen Research. All other authors report no biomedical financial interests or potential conflicts of interest.
Figures

Update of
-
Antidepressant Exposure and DNA Methylation: Insights from a Methylome-Wide Association Study.medRxiv [Preprint]. 2024 May 3:2024.05.01.24306640. doi: 10.1101/2024.05.01.24306640. medRxiv. 2024. Update in: Nat Commun. 2025 Feb 24;16(1):1908. doi: 10.1038/s41467-024-55356-x. PMID: 38746357 Free PMC article. Updated. Preprint.
References
-
- GBD 2017 Dis Injury Incidence Pr, James, S. L. G. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet. 2018;392:1789-1858. 10.1016/S0140-6736(18)32279-7. - PMC - PubMed
-
- Souery, D. et al. Switching antidepressant class does not improve response or remission in treatment-resistant depression. J. Clin. Psychopharmacol.31, 512–516 (2011). - PubMed
MeSH terms
Substances
Grants and funding
- 264146/Academy of Finland (Suomen Akatemia)
- ES/S012567/1/RCUK | Economic and Social Research Council (ESRC)
- 1113400/Department of Health | National Health and Medical Research Council (NHMRC)
- 480-15-001/674/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research)
- EP-C-15-001/EPA/EPA/United States
- EP/S02431X/1/RCUK | Medical Research Council (MRC)
- 221890/Z/20/Z/Royal Society
- Disconnected Mind project/Age UK
- R01 AA012502/AA/NIAAA NIH HHS/United States
- 308248/Academy of Finland (Suomen Akatemia)
- 141054/Academy of Finland (Suomen Akatemia)
- 227063/Z/23/Z/Wellcome Trust (Wellcome)
- 220857/Z/20/Z/Wellcome Trust (Wellcome)
- 118555/Academy of Finland (Suomen Akatemia)
- 251316/Academy of Finland (Suomen Akatemia)
- R01 HD077482/HD/NICHD NIH HHS/United States
- [Disconnected Mind project]/Age UK
- 1173790/Department of Health | National Health and Medical Research Council (NHMRC)
- 328685/Academy of Finland (Suomen Akatemia)
- 847776/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 307339/Academy of Finland (Suomen Akatemia)
- 263278/Academy of Finland (Suomen Akatemia)
- R01 AA009203/AA/NIAAA NIH HHS/United States
- R01 HL104125/HL/NHLBI NIH HHS/United States
- BB/W008793/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- #1105/Sigrid Juséliuksen Säätiö (Sigrid Jusélius Foundation)
- K02 AA018755/AA/NIAAA NIH HHS/United States
- MR/R024065/1/RCUK | Medical Research Council (MRC)
- R01 MH099110/MH/NIMH NIH HHS/United States
- 100499/Academy of Finland (Suomen Akatemia)
- K05 AA000145/AA/NIAAA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AA015416/AA/NIAAA NIH HHS/United States
- 1057/Sigrid Juséliuksen Säätiö (Sigrid Jusélius Foundation)
- 205585/Academy of Finland (Suomen Akatemia)
- 226770/Z/22/Z/Wellcome Trust (Wellcome)
- U01 MH109528/MH/NIMH NIH HHS/United States
- R37 AA012502/AA/NIAAA NIH HHS/United States
- R01MH124981/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01 MH124981/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

