NIEAs elicited by wild-type SARS-CoV-2 primary infection fail to enhance the infectivity of Omicron variants
- PMID: 39994733
- PMCID: PMC11849304
- DOI: 10.1186/s12985-025-02667-0
NIEAs elicited by wild-type SARS-CoV-2 primary infection fail to enhance the infectivity of Omicron variants
Abstract
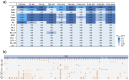
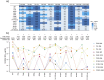
SARS-CoV-2 infection widely induces antibody response targeting diverse viral proteins, including typical representative N-terminal domain (NTD), receptor-binding domain (RBD), and S2 subunit of spike. A lot of NTD-, RBD-, and S2-specific monoclonal antibodies (mAbs) have been isolated from COVID-19 convalescents, some of which displaying potent activities to inhibit viral infection. However, a small portion of NTD-specific mAbs elicited by wild-type (WT) SARS-CoV-2 primary infection could facilitate the virus entry into target cells in vitro, so called NTD-targeting infection-enhancing antibodies (NIEAs). To date, SARS-CoV-2 has evolved to massive variants carrying various NTD mutations, especially recent Omicron BA.2.86 and JN.1. In this study, we investigated whether these WT-NIEAs could still enhance the infectivity of emerging Omicron variants. Nine novel WT-NIEAs with diverse germline gene usage were identified from 3 individuals, effectively enlarging available antibody panel of NIEAs. Bivalent binding of NIEAs to inter-spike contributed to their infection-enhancing activities. WT-NIEAs could enhance the infectivity of SARS-CoV-2 variants emerged before Omicron, but ineffective to Omicron variants including BA.2.86 and JN.1, which was because of their changed antigenicity of NTDs. Overall, these data clearly demonstrated the cross-reactivity of these pre-existed WT-NIEAs to a series of SARS-CoV-2 variants, helping to evaluate the risk of enhanced infection of emerging variants in future.
Keywords: Cross-reactivity; Infection-enhancing activity; NIEAs; NTD; SARS-CoV-2 variants.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Ethics Committee of Shenzhen Third People’s Hospital, China (approval number: 2021-030). The biological sample bank of the Shenzhen Third People’s Hospital supplied the participant’s samples and clinical information. Consent for publication: All authors approved the submission of the manuscript for publication. Competing interests: The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

