Evidence of blood-brain barrier dysfunction and CSF immunoglobulin synthesis in Down Syndrome Regression Disorder
- PMID: 39996411
- PMCID: PMC12040517
- DOI: 10.1002/acn3.52299
Evidence of blood-brain barrier dysfunction and CSF immunoglobulin synthesis in Down Syndrome Regression Disorder
Abstract
Objectives: This study sought to evaluate proteomic, metabolomic, and immune signatures in the cerebrospinal fluid of individuals with Down Syndrome Regression Disorder (DSRD).
Methods: A prospective case-control study comparing proteomic, metabolomic, and immune profiles in individuals with DSRD was performed. Samples were obtained from a biorepository of affected individuals and compared to clinically available data and previously obtained neurodiagnostic studies. Individuals with DSRD were compared to individuals with established neuroinflammatory conditions (e.g., multiple sclerosis), and neurotypical controls undergoing a lumbar puncture for headaches. Samples underwent high-throughput proteomic, metabolomic, and immune marker profiling. Data was compared across groups and clinical phenotypes. Gene set enrichment analysis and pathway analyses were utilized to analyze the data.
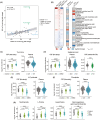
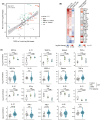
Results: In total, 34 individuals with DSRD, 22 neuroinflammatory controls, and 27 neurotypical controls were enrolled in the study. We observed a highly significant concordance in dysregulated proteomics signatures in DSRD and neuroinflammatory controls versus healthy controls, most prominently upregulation of many immunoglobulin sequences. In addition, individuals with DSRD displayed strong upregulation of liver-derived plasma proteins and erythrocyte proteins in the CSF, indicating poor blood-brain barrier integrity. The immune marker profile of DSRD is clearly similar to other neuroimmunological conditions, including strong elevation of MIP3-α, eotaxin, and IFN-γ.
Interpretation: Individuals with DSRD have unique CSF proteomic and metabolomic signatures consistent with neuroinflammation and increased blood-brain barrier permeability. The CSF of individuals with DSRD was more comparable to individuals with neuroinflammatory disorders than neurotypical controls, indicating the potential for an immune etiology of disease.
© 2025 The Author(s). Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
The authors report no relevant conflicts of interest‐related data presented.
Figures


References
-
- de Graaf G, Buckley F, Skotko BG. Estimates of the live births, natural losses, and elective terminations with Down syndrome in the United States. Am J Med Genet A. 2015;167:756‐767. - PubMed
-
- Rosso M, Fremion E, Santoro SL, et al. Down syndrome disintegrative disorder: a clinical regression syndrome of increasing importance. Pediatrics. 2020;145:e20192939. - PubMed
-
- Worley G, Crissman BG, Cadogan E, Milleson C, Adkins DW, Kishnani PS. Down syndrome disintegrative disorder: new‐onset autistic regression, dementia, and insomnia in older children and adolescents with Down syndrome. J Child Neurol. 2015;30:1147‐1152. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
