Lipid Droplets Metabolism Mediated by ANXA7-PPARγ Signaling Axis Regulates Spinal Cord Injury Repair in Mice
- PMID: 39996504
- PMCID: PMC12021101
- DOI: 10.1002/advs.202417326
Lipid Droplets Metabolism Mediated by ANXA7-PPARγ Signaling Axis Regulates Spinal Cord Injury Repair in Mice
Abstract
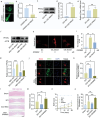
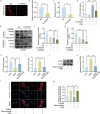
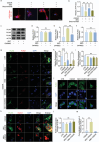
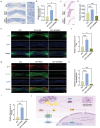
Spinal cord injury is characterized by high incidence and high disability, and the specific targets and drugs have not yet been explored. Lipid droplet is a type of organelles that regulates lipid metabolism and oxidative stress. And the regulatory mechanisms of lipid droplets on spinal cord injury remain unclear. Herein, it is found that GTPase activation of Annexin A7 (ANXA7) promotes the up-regulation of genes related to lipid droplet formation. ANXA7 can interact with peroxisome proliferator-activated receptor gamma (PPARγ) to enhance the stability of PPARγ, and promote lipid droplet formation and interaction with mitochondria through promoting Perilipin 5 expression. Then, oxidative stress and lipid peroxidation are inhibited due to the promotion of nuclear factor erythroid 2-related factor 2 (NRF2) nuclear translocation and expression of glutathione peroxidase 4 (GPX4). ANXA7 activation promotes lipid droplet formation and mitochondria-lipid droplet interaction by enhancing nuclear translocation of PPARγ, which contributes to inhibiting lipid peroxidation and neuron damage. Furthermore, activation of PPARγ can promote neural function recovery and spinal cord repair in mice. The focus of this study is to investigate the effects of lipid droplets regulated by ANXA7/PPARγ, providing new targets and strategies for spinal cord injury.
Keywords: ANXA7; PPARγ; lipid droplet; oxidative stress; spinal cord injury.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Mechanism of Mettl14 regulating AIM2 inflammasome activation and neuronal apoptosis and pyroptosis in spinal cord injury by mediating PPARγ m6A methylation.J Physiol Biochem. 2024 Nov;80(4):881-894. doi: 10.1007/s13105-024-01047-6. Epub 2024 Oct 14. J Physiol Biochem. 2024. PMID: 39400644
-
Annexin A7 deficiency potentiates cardiac NFAT activity promoting hypertrophic signaling.Biochem Biophys Res Commun. 2014 Feb 28;445(1):244-9. doi: 10.1016/j.bbrc.2014.01.186. Epub 2014 Feb 6. Biochem Biophys Res Commun. 2014. PMID: 24508799
-
Effect of Rab18 on liver injury and lipid accumulation by regulating perilipin 2 and peroxisome proliferator-activated receptor gamma in non-alcoholic fatty liver disease.J Gastroenterol Hepatol. 2024 Oct;39(10):2219-2227. doi: 10.1111/jgh.16676. Epub 2024 Jul 18. J Gastroenterol Hepatol. 2024. PMID: 39030773
-
The role of Nrf2 and PPARgamma in the improvement of oxidative stress in hypertension and cardiovascular diseases.Physiol Res. 2020 Dec 31;69(Suppl 4):S541-S553. doi: 10.33549/physiolres.934612. Physiol Res. 2020. PMID: 33656904 Free PMC article. Review.
-
ANXA7-GTPase as Tumor Suppressor: Mechanisms and Therapeutic Opportunities.Methods Mol Biol. 2017;1513:23-35. doi: 10.1007/978-1-4939-6539-7_3. Methods Mol Biol. 2017. PMID: 27807828 Review.
References
-
- Collaborators G. N., Lancet. Neurol. 2019, 18, 459. - PubMed
-
- Sofroniew M. V., Nature 2018, 557, 343. - PubMed
-
- Rowald A., Komi S., Demesmaeker R., Baaklini E., Hernandez‐Charpak S. D., Paoles E., Montanaro H., Cassara A., Becce F., Lloyd B., Newton T., Ravier J., Kinany N., D'Ercole M., Paley A., Hankov N., Varescon C., McCracken L., Vat M., Caban M., Watrin A., Jacquet C., Bole‐Feysot L., Harte C., Lorach H., Galvez A., Tschopp M., Herrmann N., Wacker M., Geernaert L., et al., Nat. Med. 2022, 28, 260. - PubMed
-
- David G., Mohammadi S., Martin A. R., Cohen‐Adad J., Weiskopf N., Thompson A., Freund P., Nat. Rev. Neurol. 2019, 15, 718. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
