Rescuing DNMT1 fails to fully reverse the molecular and functional repercussions of its loss in mouse embryonic stem cells
- PMID: 39997223
- PMCID: PMC11851107
- DOI: 10.1093/nar/gkaf130
Rescuing DNMT1 fails to fully reverse the molecular and functional repercussions of its loss in mouse embryonic stem cells
Abstract
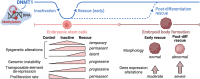
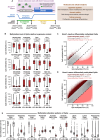
Epigenetic mechanisms are crucial for developmental programming and can be disrupted by environmental stressors, increasing susceptibility to disease. This has sparked interest in therapies for restoring epigenetic balance, but it remains uncertain whether disordered epigenetic mechanisms can be fully corrected. Disruption of DNA methyltransferase 1 (DNMT1), responsible for DNA methylation maintenance, has particularly devastating biological consequences. Therefore, here we explored if rescuing DNMT1 activity is sufficient to reverse the effects of its loss utilizing mouse embryonic stem cells. However, only partial reversal could be achieved. Extensive changes in DNA methylation, histone modifications, and gene expression were detected, along with transposable element derepression and genomic instability. Reduction of cellular size, complexity, and proliferation rate were observed, as well as lasting effects in germ layer lineages and embryoid bodies. Interestingly, by analyzing the impact on imprinted regions, we uncovered 20 regions exhibiting imprinted-like signatures. Notably, while many permanent effects persisted throughout Dnmt1 inactivation and rescue, others arose from the rescue intervention. Lastly, rescuing DNMT1 after differentiation initiation worsened outcomes, reinforcing the need for early intervention. Our findings highlight the far-reaching functions of DNMT1 and provide valuable perspectives on the repercussions of epigenetic perturbations during early development and the challenges of rescue interventions.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

