Deep Learning-Based Molecular Fingerprint Prediction for Metabolite Annotation
- PMID: 39997757
- PMCID: PMC11857613
- DOI: 10.3390/metabo15020132
Deep Learning-Based Molecular Fingerprint Prediction for Metabolite Annotation
Abstract
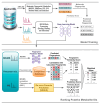
Background/Objectives: Liquid chromatography coupled with mass spectrometry (LC-MS) is a commonly used platform for many metabolomics studies. However, metabolite annotation has been a major bottleneck in these studies in part due to the limited publicly available spectral libraries, which consist of tandem mass spectrometry (MS/MS) data acquired from just a fraction of known compounds. Application of deep learning methods is increasingly reported as an alternative to spectral matching due to their ability to map complex relationships between molecular fingerprints and mass spectrometric measurements. The objectives of this study are to investigate deep learning methods for molecular fingerprint based on MS/MS spectra and to rank putative metabolite IDs according to similarity of their known and predicted molecular fingerprints. Methods: We trained three types of deep learning methods to model the relationships between molecular fingerprints and MS/MS spectra. Prior to training, various data processing steps, including scaling, binning, and filtering, were performed on MS/MS spectra obtained from National Institute of Standards and Technology (NIST), MassBank of North America (MoNA), and Human Metabolome Database (HMDB). Furthermore, selection of the most relevant m/z bins and molecular fingerprints was conducted. The trained deep learning models were evaluated on ranking putative metabolite IDs obtained from a compound database for the challenges in Critical Assessment of Small Molecule Identification (CASMI) 2016, CASMI 2017, and CASMI 2022 benchmark datasets. Results: Feature selection methods effectively reduced redundant molecular and spectral features prior to model training. Deep learning methods trained with the truncated features have shown comparable performances against CSI:FingerID on ranking putative metabolite IDs. Conclusion: The results demonstrate a promising potential of deep learning methods for metabolite annotation.
Keywords: LC-MS/MS; deep learning; metabolite identification; molecular fingerprint prediction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Convolutional Neural Network-Based Compound Fingerprint Prediction for Metabolite Annotation.Metabolites. 2022 Jun 29;12(7):605. doi: 10.3390/metabo12070605. Metabolites. 2022. PMID: 35888729 Free PMC article.
-
Deep Learning Based Metabolite Annotation.Annu Int Conf IEEE Eng Med Biol Soc. 2023 Jul;2023:1-4. doi: 10.1109/EMBC40787.2023.10341007. Annu Int Conf IEEE Eng Med Biol Soc. 2023. PMID: 38082953 Free PMC article.
-
MetFID: artificial neural network-based compound fingerprint prediction for metabolite annotation.Metabolomics. 2020 Sep 30;16(10):104. doi: 10.1007/s11306-020-01726-7. Metabolomics. 2020. PMID: 32997169 Free PMC article.
-
Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics.Metabolites. 2018 May 10;8(2):31. doi: 10.3390/metabo8020031. Metabolites. 2018. PMID: 29748461 Free PMC article. Review.
-
Insights into predicting small molecule retention times in liquid chromatography using deep learning.J Cheminform. 2024 Oct 7;16(1):113. doi: 10.1186/s13321-024-00905-1. J Cheminform. 2024. PMID: 39375739 Free PMC article. Review.
References
-
- NIST/EPA/NIH Mass Spectral Library. [(accessed on 22 July 2024)]; Available online: http://www.nist.gov/srd/nist1a.cfm.
Grants and funding
LinkOut - more resources
Full Text Sources

