Neurotoxins Acting on TRPV1-Building a Molecular Template for the Study of Pain and Thermal Dysfunctions
- PMID: 39998081
- PMCID: PMC11861614
- DOI: 10.3390/toxins17020064
Neurotoxins Acting on TRPV1-Building a Molecular Template for the Study of Pain and Thermal Dysfunctions
Abstract
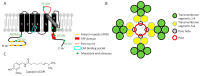
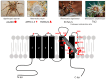
Transient Receptor Potential (TRP) channels are ubiquitous proteins involved in a wide range of physiological functions. Some of them are expressed in nociceptors and play a major role in the transduction of painful stimuli of mechanical, thermal, or chemical origin. They have been described in both human and rodent systems. Among them, TRPV1 is a polymodal channel permeable to cations, with a highly conserved sequence throughout species and a homotetrameric structure. It is sensitive to temperature above 43 °C and to pH below 6 and involved in various functions such as thermoregulation, metabolism, and inflammatory pain. Several TRPV1 mutations have been associated with human channelopathies related to pain sensitivity or thermoregulation. TRPV1 is expressed in a large part of the peripheral and central nervous system, most notably in sensory C and Aδ fibers innervating the skin and internal organs. In this review, we discuss how the transduction of nociceptive messages is activated or impaired by natural compounds and peptides targeting TRPV1. From a pharmacological point of view, capsaicin-the spicy ingredient of chilli pepper-was the first agonist described to activate TRPV1, followed by numerous other natural molecules such as neurotoxins present in plants, microorganisms, and venomous animals. Paralleling their adaptive protective benefit and allowing venomous species to cause acute pain to repel or neutralize opponents, these toxins are very useful for characterizing sensory functions. They also provide crucial tools for understanding TRPV1 functions from a structural and pharmacological point of view as this channel has emerged as a potential therapeutic target in pain management. Therefore, the pharmacological characterization of TRPV1 using natural toxins is of key importance in the field of pain physiology and thermal regulation.
Keywords: TRPV1; agonists; antagonists; pain; thermoregulation; toxins; venoms.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

 ), [92] for DkTx (●), [99] for BmP01 (
), [92] for DkTx (●), [99] for BmP01 ( ), [100,101] for RhTx (♦), and [102] for HNTX-XXI (
), [100,101] for RhTx (♦), and [102] for HNTX-XXI ( ) and HNTX-XXII (
) and HNTX-XXII ( ). Latoia consocia is from ©Daniel Ruyle. All other images are from Wikimedia commons.
). Latoia consocia is from ©Daniel Ruyle. All other images are from Wikimedia commons.
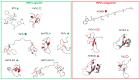
 ), and [110] for HCRG21 (▲). All images are from Wikimedia commons.
), and [110] for HCRG21 (▲). All images are from Wikimedia commons.
References
-
- Raja S.N., Carr D.B., Cohen M., Finnerup N.B., Flor H., Gibson S., Keefe F.J., Mogil J.S., Ringkamp M., Sluka K.A., et al. The Revised International Association for the Study of Pain Definition of Pain: Concepts, Challenges, and Compromises. Pain. 2020;161:1976–1982. doi: 10.1097/j.pain.0000000000001939. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

