SARS-CoV-2 infectivity can be modulated through bacterial grooming of the glycocalyx
- PMID: 39998226
- PMCID: PMC11980591
- DOI: 10.1128/mbio.04015-24
SARS-CoV-2 infectivity can be modulated through bacterial grooming of the glycocalyx
Abstract
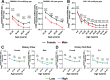
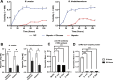
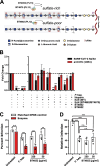
The gastrointestinal (GI) tract is a site of replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and GI symptoms are often reported by patients. SARS-CoV-2 cell entry depends upon heparan sulfate (HS) proteoglycans, which commensal bacteria that bathe the human mucosa are known to modify. To explore human gut HS-modifying bacterial abundances and how their presence may impact SARS-CoV-2 infection, we developed a task-based analysis of proteoglycan degradation on large-scale shotgun metagenomic data. We observed that gut bacteria with high predicted catabolic capacity for HS differ by age and sex, factors associated with coronavirus disease 2019 (COVID-19) severity, and directly by disease severity during/after infection, but do not vary between subjects with COVID-19 comorbidities or by diet. Gut commensal bacterial HS-modifying enzymes reduce spike protein binding and infection of authentic SARS-CoV-2, suggesting that bacterial grooming of the GI mucosa may impact viral susceptibility.IMPORTANCESevere acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for coronavirus disease 2019, can infect the gastrointestinal (GI) tract, and individuals who exhibit GI symptoms often have more severe disease. The GI tract's glycocalyx, a component of the mucosa covering the large intestine, plays a key role in viral entry by binding SARS-CoV-2's spike protein via heparan sulfate (HS). Here, using metabolic task analysis of multiple large microbiome sequencing data sets of the human gut microbiome, we identify a key commensal human intestinal bacteria capable of grooming glycocalyx HS and modulating SARS-CoV-2 infectivity in vitro. Moreover, we engineered the common probiotic Escherichia coli Nissle 1917 (EcN) to effectively block SARS-CoV-2 binding and infection of human cell cultures. Understanding these microbial interactions could lead to better risk assessments and novel therapies targeting viral entry mechanisms.
Keywords: Covid; Heparan Sulfate; SARS-CoV-2; aging; human microbiome.
Conflict of interest statement
The authors declare a conflict of interest (see Acknowledgments).
Figures

References
-
- Elshazli RM, Kline A, Elgaml A, Aboutaleb MH, Salim MM, Omar M, Munshi R, Mankowski N, Hussein MH, Attia AS, Toraih EA, Settin A, Killackey M, Fawzy MS, Kandil E. 2021. Gastroenterology manifestations and COVID-19 outcomes: a meta-analysis of 25,252 cohorts among the first and second waves. J Med Virol 93:2740–2768. doi:10.1002/jmv.26836 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01 GM069811/GM/NIGMS NIH HHS/United States
- NNF20SA0066621/Novo Nordisk Foundation
- R01 ES027595/ES/NIEHS NIH HHS/United States
- 1P30DK120515/HHS | National Institutes of Health (NIH)
- U19 AI116497/AI/NIAID NIH HHS/United States
- R00RG2503/UC | University of California, San Diego (UCSD)
- 2031989/National Science Foundation (NSF)
- ANR-17-CE11-0014/French National Research Agency
- 335525/Academy of Finland
- 3022/Emerald Foundation
- 1DP1AT010885/HHS | National Institutes of Health (NIH)
- U01 CA199277/CA/NCI NIH HHS/United States
- R01-CA077398/HHS | National Institutes of Health (NIH)
- R01ES027595/HHS | National Institutes of Health (NIH)
- UM1 CA164917/CA/NCI NIH HHS/United States
- R01GM069811/HHS | National Institutes of Health (NIH)
- RG/18/13/33946/British Heart Foundation (BHF)
- 2018-72190270/ANID Becas Chile Doctorado
- Emil Aaltosen Säätiö (Emil Aaltonen Foundation)
- UH2AI153029/HHS | National Institutes of Health (NIH)
- Health Data Research UK (HDR UK)
- AI Horizons Network/IBM | IBM Research
- P30-CA023100/HHS | National Institutes of Health (NIH)
- P30-CA033572/HHS | National Institutes of Health (NIH)
- DP1 AT010885/AT/NCCIH NIH HHS/United States
- NIHR | NIHR Cambridge Biomedical Research Centre (NIHR Cambridge BRC)
- Sydäntutkimussäätiö (Finnish Foundation for Cardiovascular Research)
- UH2 AI153029/AI/NIAID NIH HHS/United States
- P30 DK120515/DK/NIDDK NIH HHS/United States
- 101046041/Union's Horizon Europe Research and Innovation 459 Actions
- Munz Chair of Cardiovascular Prediction and Prevention
- 321351 and 354447/Academy of Finland
- R35GM119850/HHS | National Institutes of Health (NIH)
- UM1-CA164917/HHS | National Institutes of Health (NIH)
- P30 CA023100/CA/NCI NIH HHS/United States
- R01 CA077398/CA/NCI NIH HHS/United States
- BRC-1215-20014/NIHR | NIHR Cambridge Biomedical Research Centre (NIHR Cambridge BRC)
- R35 GM119850/GM/NIGMS NIH HHS/United States
- RG/13/13/30194/British Heart Foundation (BHF)
- P30 CA033572/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 2038509/National Science Foundation (NSF)
- N/A/UC San Diego Center for Microbiome Innovation
- P01 HL131474/HL/NHLBI NIH HHS/United States
- U01-CA199277/HHS | National Institutes of Health (NIH)
- 321356/Academy of Finland
- Alfred Benzon Foundation (The Alfred Benzon Foundation)
- ANR-20-CE44-0005/French National Research Agency
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
