Characterization of vaginal microbiomes in clinician-collected bacterial vaginosis diagnosed samples
- PMID: 39998243
- PMCID: PMC11960135
- DOI: 10.1128/spectrum.02582-24
Characterization of vaginal microbiomes in clinician-collected bacterial vaginosis diagnosed samples
Abstract
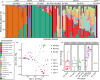
Bacterial vaginosis (BV) is a type of vaginal inflammation caused by bacterial overgrowth, upsetting the healthy microbiome of the vagina. Existing clinical testing for BV is primarily based upon physical and microscopic examination of vaginal secretions. Modern PCR-based clinical tests target panels of BV-associated microbes, such as the Labcorp NuSwab test that targets Atopobium (Fannyhessea) vaginae, Megasphaera-1, and Bacterial Vaginosis Associated Bacterium (BVAB)-2. Remnant clinician-collected NuSwab vaginal swabs underwent DNA extraction and 16S V3-V4 rRNA gene sequencing to profile microbes in addition to those included in the Labcorp NuSwab test. Community state types (CSTs) were determined using the most abundant taxon detected in each sample. PCR results for NuSwab panel microbial targets were compared against the corresponding microbiome profiles. Metabolic pathway abundances were characterized via metagenomic prediction from amplicon sequence variants (ASVs). 16S V3-V4 rRNA gene sequencing of 75 remnant vaginal swabs yielded 492 unique 16S V3-V4 ASVs, identifying 83 unique genera. NuSwab microbe quantification was strongly concordant with quantification by sequencing (P < 0.01). Samples in CST-I (18 of 18, 100%), CST-II (three of three, 100%), CST-III (15 of 17, 88%), and CST-V (one of one, 100%) were largely categorized as BV-negative via the NuSwab panel, while most CST-IV samples (28 of 36, 78%) were BV-positive or BV-indeterminate. BV-associated microbial and predicted metabolic signatures were shared across multiple CSTs. These findings highlight robust sequencing-based quantification of Labcorp NuSwab BV microbes, accurate discrimination of vaginal microbiome CSTs dominated by distinct Lactobacilli, and expanded the identification of BV-associated bacterial and metabolic biomarkers.IMPORTANCEBacterial vaginosis (BV) poses a significant health burden for women during reproductive years and onward. Current BV diagnostics rely on either panels of select microbes or on physical and microscopic evaluations by technicians. Here, we sequenced the microbiome profiles of samples previously diagnosed by the Labcorp NuSwab test to better understand disruptions to the vaginal microbiome during BV. We show that microbial sequencing can faithfully reproduce targeted PCR diagnostic results and can improve our knowledge of healthy and BV-associated microbial and metabolic biomarkers. This work highlights a robust, agnostic BV classification scheme with potential for future development of sequencing-based BV diagnostic tools.
Keywords: 16S rRNA; bacterial vaginosis; vaginal microbiome.
Conflict of interest statement
All authors are current or former employees of Labcorp, a provider of clinical diagnostic services.
Figures




References
-
- Kuruvilla S, Bustreo F, Kuo T, Mishra CK, Taylor K, Fogstad H, Gupta GR, Gilmore K, Temmerman M, Thomas J, et al. . 2016. The Global strategy for women’s, children’s and adolescents’ health (2016-2030): a roadmap based on evidence and country experience. Bull World Health Organ 94:398–400. doi:10.2471/BLT.16.170431 - DOI - PMC - PubMed
-
- Joseph RJ, Ser H-L, Kuai Y-H, Tan L-H, Arasoo VJT, Letchumanan V, Wang L, Pusparajah P, Goh B-H, Ab Mutalib N-S, Chan K-G, Lee L-H. 2021. Finding a balance in the vaginal microbiome: how do we treat and prevent the occurrence of bacterial vaginosis? Antibiotics (Basel) 10:719. doi:10.3390/antibiotics10060719 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

