A strain of Streptococcus mitis inhibits biofilm formation of caries pathogens via abundant hydrogen peroxide production
- PMID: 39998256
- PMCID: PMC11921374
- DOI: 10.1128/aem.02192-24
A strain of Streptococcus mitis inhibits biofilm formation of caries pathogens via abundant hydrogen peroxide production
Abstract
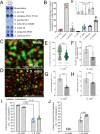
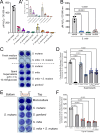
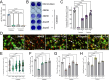
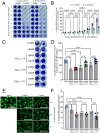
Commensal oral streptococci that colonize supragingival biofilms deploy mechanisms to combat competitors within their niche. Here, we determined that Streptococcus mitis more effectively inhibited biofilm formation of Streptococcus mutans compared to other oral streptococci. This phenotype was common among all isolates of S. mutans, but was specific to a single strain of S. mitis, ATCC 49456. We documented ATCC 49456 to accumulate four to five times more hydrogen peroxide (H2O2) than other Streptococcus species tested, and 5-18 times more than other S. mitis strains assayed. S. mutans biofilm formation inhibition was dependent on cell contact/proximity and reduced when grown in media containing catalase or with a S. mitis mutant of pyruvate oxidase (spxB; pox), confirming that SpxB-dependent H2O2 production was a major antagonistic factor. Addition of S. mitis within hours after S. mutans inoculation was effective at reducing biofilm biomass, but not for 24 h pre-formed biofilms in an SpxB-dependent manner. Transcriptome analysis revealed responses for both S. mitis and S. mutans, with several S. mutans differentially expressed genes following a gene expression pattern we have previously described, while others being unique to the interaction with S. mitis. Finally, we show that S. mitis also affected coculture biofilm formation of several other commensal streptococci as well as cariogenic Streptococcus sobrinus. Our study shows that strains with abundant H2O2 production are effective at inhibiting initial growth of caries pathogens like S. mutans, but are less effective at disrupting pre-formed biofilms and have the potential to influence the stability of other oral commensal strains.IMPORTANCEAntagonistic properties displayed by oral bacteria have been sought as therapeutic approaches against dental caries pathogens like Streptococcus mutans. An emergent theme has been the ability of select strains that produce high amounts of hydrogen peroxide to effectively inhibit the growth of S. mutans within in vitro and in vivo models. Our study builds on these previous findings by determining that Streptococcus mitis ATCC 49456 is a high hydrogen peroxide producer, compared to other Streptococcus species as well as additional strains of S. mitis. In addition to S. mutans, we show that ATCC 49456 also affects biofilm formation of other oral streptococci, a non-desirable trait that should be weighed heavily for strains under consideration as probiotics. Further phenotypic characterization of strains like S. mitis ATCC 49456 in mixed-species settings will allow us to hone in on qualities that are optimal for probiotic strains that are intended to prevent the emergence of odontopathogens.
Keywords: biofilms; dental caries; hydrogen peroxide; intermicrobial interactions; oral biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Update of
-
A Strain of Streptococcus mitis Inhibits Biofilm Formation of Caries Pathogens via Abundant Hydrogen Peroxide Production.bioRxiv [Preprint]. 2024 Aug 6:2024.08.06.606862. doi: 10.1101/2024.08.06.606862. bioRxiv. 2024. Update in: Appl Environ Microbiol. 2025 Mar 19;91(3):e0219224. doi: 10.1128/aem.02192-24. PMID: 39149263 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

