Multiplexed epigenetic memory editing using CRISPRoff sensitizes glioblastoma to chemotherapy
- PMID: 39998382
- PMCID: PMC12309708
- DOI: 10.1093/neuonc/noaf055
Multiplexed epigenetic memory editing using CRISPRoff sensitizes glioblastoma to chemotherapy
Abstract
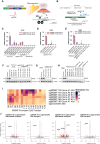
Background: Glioblastoma (GBM) carries a poor prognosis, and new therapeutic strategies are necessary to improve outcomes for patients with this disease. Alkylating chemotherapies including temozolomide (TMZ) and lomustine (CCNU) are critical for treating GBM, but resistance mechanisms, including hypomethylation of O6-methylguanine-DNA methyltransferase (MGMT) promoter, undermine treatment. CRISPRoff is a programmable epigenetic memory editor that can induce stable and heritable gene silencing after transient delivery, and we hypothesize that CRISPRoff could potentiate the activity of TMZ and CCNU through long-term suppression of target genes.
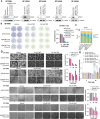
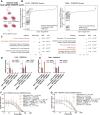
Methods: We transiently delivered CRISPRoff mRNA along with sgRNAs against target genes using both electroporation and lipid nanoparticles (LNPs) into established GBM cell lines, patient-derived primary GBM cultures, and orthotopic GBM xenografts. Gene repression, specificity, and stability were measured by RT-qPCR, Western blot, bisulfite sequencing, and RNA sequencing. Sensitivity to chemotherapies was measured by cell viability dose-response, microscopy, and bioluminescence imaging. Genome-wide mapping of CCNU sensitizers was performed using CRISPRi screens.
Results: CRISPRoff induced complete suppression of MGMT and sensitization to TMZ that was stable for over 8 months of continuous cell propagation. GBM orthotopic tumors treated with CRISPRoff against MGMT demonstrated sensitivity to TMZ in vivo, and CRISPRoff delivery resulted in chemosensitivity in patient-derived primary GBM. Genome-wide CRISPRi screens identified combinatorial genetic vulnerabilities (BRIP1, FANCE) that were targetable by multiplexed CRISPRoff to achieve sensitization to CCNU.
Conclusion: Transient delivery of a site-specific epigenetic memory can induce stable, complete, and multiplexed suppression of target genes for therapeutic application in GBM.
Keywords: CCNU; CRISPR; epigenetic editing; glioblastoma; temozolomide.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Conflict of interest statement
L.A.G has filed patents on CRISPR tools and CRISPR functional genomics. L.A.G. is a co-founder of Chroma Medicine and serves on the scientific advisory board of Myllia Biotechnology. A.M. is a cofounder of Site Tx, Arsenal Biosciences, Spotlight Therapeutics and Survey Genomics, serves on the boards of directors at Site Tx, Spotlight Therapeutics and Survey Genomics, is a member of the scientific advisory boards of Site Tx, Arsenal Biosciences, Cellanome, Spotlight Therapeutics, Survey Genomics, NewLimit, Amgen, and Tenaya, owns stock in Arsenal Biosciences, Site Tx, Cellanome, Spotlight Therapeutics, NewLimit, Survey Genomics, Tenaya and Lightcast and has received fees from Site Tx, Arsenal Biosciences, Cellanome, Spotlight Therapeutics, NewLimit, Gilead, Pfizer, 23andMe, PACT Pharma, Juno Therapeutics, Tenaya, Lightcast, Trizell, Vertex, Merck, Amgen, Genentech, GLG, ClearView Healthcare, AlphaSights, Rupert Case Management, Bernstein and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ. The Marson laboratory has received research support from the Parker Institute for Cancer Immunotherapy, the Emerson Collective, Arc Institute, Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead and Anthem and reagents from Genscript and Illumina.
Figures



References
-
- Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. - PubMed
-
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

