CLIC5 promotes myoblast differentiation and skeletal muscle regeneration via the BGN-mediated canonical Wnt/β-catenin signaling pathway
- PMID: 39999205
- PMCID: PMC11468980
- DOI: 10.1126/sciadv.adq6795
CLIC5 promotes myoblast differentiation and skeletal muscle regeneration via the BGN-mediated canonical Wnt/β-catenin signaling pathway
Abstract
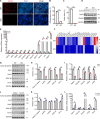
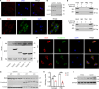
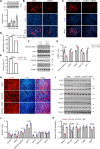
Myoblast differentiation plays a vital role in skeletal muscle regeneration. However, the protein-coding genes controlling this process remain incompletely understood. Here, we showed that chloride intracellular channel 5 (CLIC5) exerts a critical role in mediating myogenesis and skeletal muscle regeneration. Deletion of CLIC5 in skeletal muscle leads to reduced muscle weight and decreases the number and differentiation potential of satellite cells. In vitro, CLIC5 consistently inhibits myoblast proliferation while promoting myotube formation. CLIC5 promotes myogenic differentiation by activating the canonical Wnt/β-catenin signaling pathway in a biglycan (BGN)-dependent manner. CLIC5 deletion impairs muscle regeneration. Paired box gene 7 (Pax7) expression and the activity of BGN-mediated canonical Wnt/β-catenin signaling are reduced in CLIC5-deficient mice. Conversely, increasing CLIC5 levels in skeletal muscles enhances muscle regeneration capacity. In conclusion, our findings underscore CLIC5 as a pivotal regulator of myogenesis and skeletal muscle regeneration, functioning through interaction with BGN to activate the canonical Wnt/β-catenin signaling pathway.
Figures




References
-
- Janssen I., Heymsfield S. B., Wang Z. M., Ross R., Skeletal muscle mass and distribution in 468 men and women aged 18–88 yr. J. Appl. Physiol. 89, 81–88 (2000). - PubMed
-
- Chargé S. B., Rudnicki M. A., Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238 (2004). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

