SeqFirst: Building equity access to a precise genetic diagnosis in critically ill newborns
- PMID: 39999847
- PMCID: PMC11947171
- DOI: 10.1016/j.ajhg.2025.02.003
SeqFirst: Building equity access to a precise genetic diagnosis in critically ill newborns
Abstract
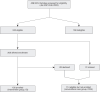
Access to a precise genetic diagnosis (PrGD) in critically ill newborns is limited and inequitable because the complex inclusion criteria used to prioritize testing eligibility omit many patients at high risk for a genetic condition. SeqFirst-neo is a program to test whether a genotype-driven workflow using simple, broad exclusion criteria to assess eligibility for rapid genome sequencing (rGS) increases access to a PrGD in critically ill newborns. All 408 newborns admitted to a neonatal intensive care unit between January 2021 and February 2022 were assessed, and of 240 eligible infants, 126 were offered rGS (i.e., intervention group [IG]) and compared to 114 infants who received conventional care in parallel (i.e., conventional care group [CCG]). A PrGD was made in 62/126 (49.2%) IG neonates compared to 11/114 (9.7%) CCG infants. The odds of receiving a PrGD were ∼9 times greater in the IG vs. the CCG, and this difference was maintained at 12 months follow-up. Access to a PrGD in the IG vs. CCG differed significantly between infants identified as non-White (34/74, 45.9% vs. 6/29, 20.7%; p = 0.024) and Black (8/10, 80.0% vs. 0/4; p = 0.015). Neonatologists were significantly less successful at predicting a PrGD in non-White than non-Hispanic White infants. The use of a standard workflow in the IG with a PrGD revealed that a PrGD would have been missed in 26/62 (42%) infants. The use of simple, broad exclusion criteria that increase access to genetic testing significantly increases access to a PrGD, improves access equity, and results in fewer missed diagnoses.
Keywords: equity; genetic diagnosis; health disparity; rapid whole-genome sequencing; rare disease.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.J.B. is on the scientific advisory board of GeneDx and has research agreements with GeneDx, Illumina, Inc., and PacBio, Inc. All GeneDx authors are/were employed by and may own stock in GeneDx, Inc. D.L.V. has served as a consultant to Illumina, Inc. D.E.M. is engaged in a research agreement with Oxford Nanopore Technologies (ONT). ONT has paid for D.E.M. to travel to speak on their behalf. D.E.M. holds stock options in MyOme.
Figures





References
-
- Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. - DOI - PMC - PubMed
-
- Cakici J.A., Dimmock D.P., Caylor S.A., Gaughran M., Clarke C., Triplett C., Clark M.M., Kingsmore S.F., Bloss C.S. A Prospective Study of Parental Perceptions of Rapid Whole-Genome and -Exome Sequencing among Seriously Ill Infants. Am. J. Hum. Genet. 2020;107:953–962. doi: 10.1016/j.ajhg.2020.10.004. - DOI - PMC - PubMed
-
- Kingsmore S.F., Cakici J.A., Clark M.M., Gaughran M., Feddock M., Batalov S., Bainbridge M.N., Carroll J., Caylor S.A., Clarke C., et al. A Randomized, Controlled Trial of the Analytic and Diagnostic Performance of Singleton and Trio, Rapid Genome and Exome Sequencing in Ill Infants. Am. J. Hum. Genet. 2019;105:719–733. doi: 10.1016/j.ajhg.2019.08.009. - DOI - PMC - PubMed
-
- Dimmock D.P., Clark M.M., Gaughran M., Cakici J.A., Caylor S.A., Clarke C., Feddock M., Chowdhury S., Salz L., Cheung C., et al. An RCT of Rapid Genomic Sequencing among Seriously Ill Infants Results in High Clinical Utility, Changes in Management, and Low Perceived Harm. Am. J. Hum. Genet. 2020;107:942–952. doi: 10.1016/j.ajhg.2020.10.003. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

