Identification of genes associated with testicular germ cell tumor susceptibility through a transcriptome-wide association study
- PMID: 39999848
- PMCID: PMC11947167
- DOI: 10.1016/j.ajhg.2025.01.022
Identification of genes associated with testicular germ cell tumor susceptibility through a transcriptome-wide association study
Abstract
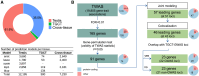
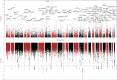
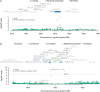
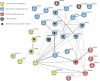
Transcriptome-wide association studies (TWASs) have the potential to identify susceptibility genes associated with testicular germ cell tumors (TGCTs). We conducted a comprehensive TGCT TWAS by integrating genome-wide association study (GWAS) summary data with predicted expression models from normal testis, TGCT tissues, and a cross-tissue panel that encompasses shared regulatory features across 22 normal tissues, including the testis. Gene associations were evaluated while accounting for variant-level effects from GWASs, followed by fine-mapping analyses in regions exhibiting multiple TWAS signals, and finally supplemented by colocalization analysis. Expression and protein patterns of identified TWAS genes were further examined in relevant tissues. Our analysis tested 19,805 gene-disease links, revealing 165 TGCT-associated genes with a false discovery rate of less than 0.01. We prioritized 46 candidate genes by considering GWAS-inflated signals, correlations between neighboring genes, and evidence of colocalization. Among these, 23 genes overlap with 22 GWAS loci, with 7 being associations not previously implicated in TGCT risk. Additionally, 23 genes located within 21 loci are at least 1 Mb away from published GWAS index variants. The 46 prioritized genes display expression levels consistent with expected expression levels in human gonadal cell types and precursor tumor cells and significant enrichment in TGCTs. Additionally, immunohistochemistry revealed protein-level accumulation of two candidate genes, ARID3B and GINM1, in both precursor and tumor cells. These findings enhance our understanding of the genetic predisposition to TGCTs and underscore the importance of further functional investigations into these candidate genes.
Keywords: colocalization analysis; testicular germ cell tumors; transcriptome-wide association study.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

