Defining the methanogenic SECIS element in vivo by targeted mutagenesis
- PMID: 40000419
- PMCID: PMC11881835
- DOI: 10.1080/15476286.2025.2472448
Defining the methanogenic SECIS element in vivo by targeted mutagenesis
Abstract
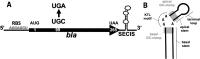
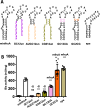
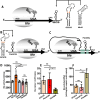
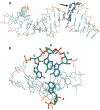
In all domains of life, Archaea, Eukarya and Bacteria, the unusual amino acid selenocysteine (Sec) is co-translationally incorporated into proteins by recoding a UGA stop codon to a sense codon. A secondary structure on the mRNA, the selenocysteine insertion sequence (SECIS), is required, but its position, secondary structure and binding partner(s) are not conserved across the tree of life. Thus far, the nature of archaeal SECIS elements has been derived mainly from sequence analyses. A recently developed in vivo reporter system was used to study the structure-function relationships of SECIS elements in Methanococcus maripaludis. Through targeted mutagenesis, we defined the minimal functional SECIS element, the parts of the SECIS where structure and not the identity of the bases are relevant for function, and identified two conserved -and invariant- adenines that are most likely to interact with the other factor(s) of the Sec recoding machinery. Finally, we demonstrated the functionality of SECIS elements in the 5`-untranslated region of the mRNA and identified a potential mechanism of SECIS repositioning in the vicinity of the UGA for efficient selenocysteine insertion.
Keywords: Archaea; Methanococcus; SECIS element; reporter; selenocysteine.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
