Establishment of an alternative splicing prognostic risk model and identification of FN1 as a potential biomarker in glioblastoma multiforme
- PMID: 40000711
- PMCID: PMC11862013
- DOI: 10.1038/s41598-025-91038-4
Establishment of an alternative splicing prognostic risk model and identification of FN1 as a potential biomarker in glioblastoma multiforme
Abstract
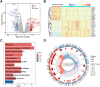
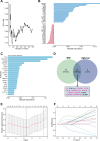
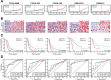
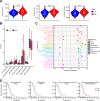
Aberrant alternative splicing and abnormal alternative splicing events (ASEs) in glioblastoma multiforme (GBM) remain largely elusive. The prognostic-associated ASEs in GBM were identified and summarized into 123 genes using GBM and LGG datasets from ASCancer Atlas and TCGA. The eleven genes (C2, COL3A1, CTSL, EIF3L, FKBP9, FN1, HPCAL1, HSPB1, IGFBP4, MANBA, PRKAR1B) were screened to develop an alternative splicing prognostic risk score (ASRS) model through machine learning algorithms. The model was trained on the TCGA-GBM cohort and validated with four external datasets from CGGA and GEO, achieving AUC values of 0.808, 0.814, 0.763, 0.859, and 0.836 for 3-year survival rates, respectively. ASRS could be an independent prognostic factor for GBM patients (HR > 1.8 across three datasets) through multivariate Cox regression analysis. The high-risk group demonstrated poorer prognosis, elevated immune scores, increased levels of immune cell infiltration, and greater differences in drug sensitivity. We found that FN1, used for model construction, contained 4 abnormal ASEs resulting in high expression of non-canonical transcripts and the presence of premature termination codon. These abnormal ASEs may be regulated by tumour-related splicing factors according to the PPI network. Furthermore, both mRNA and protein levels of FN1 were highly expressed in GBM compared to LGG, correlating with poor prognosis in GBM. In conclusion, our findings highlight the role of ASEs in affecting the progression of GBM, and the model showed a potential application for prognostic risk of patients. FN1 may serve as a promising splicing biomarker for GBM, and mechanisms of processes of aberrant splicing need to be revealed in the future.
Keywords: FN1; ASRS prognostic model; Alternative splicing; Glioblastoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: Not applicable. Consent to publish: Not applicable.
Figures



References
-
- Miller, K. D. et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J. Clin.71, 381–406 (2021). - PubMed
-
- Yang, P. et al. Management and survival rates in patients with glioma in China (2004–2010): A retrospective study from a single-institution. J. Neurooncol.113, 259–266 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- A2020177/Health Commission of Guangdong Province
- 202201010764/Guangzhou Municipal Science and Technology Bureau
- 20231219/Traditional Chinese Medicine Bureau of Guangdong Province
- "Climbing Plan" No. pdjh2023b0279/The Special Fund Project for Science and Technology Innovation Strategy of Guangdong Province
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

