Discovery and characterization of a novel telomerase alternative splicing isoform that protects lung cancer cells from chemotherapy induced cell death
- PMID: 40000722
- PMCID: PMC11861669
- DOI: 10.1038/s41598-025-90639-3
Discovery and characterization of a novel telomerase alternative splicing isoform that protects lung cancer cells from chemotherapy induced cell death
Abstract
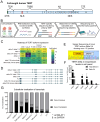
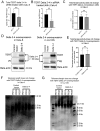
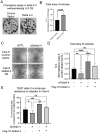
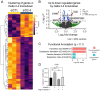
All cancer cells must adopt a telomere maintenance mechanism to achieve replicative immortality. Most human cancer cells utilize the enzyme telomerase to maintain telomeres. Alternative splicing of TERT regulates the amount and function of telomerase, however many alternative splicing isoforms of TERT have unknown functions. Single molecule long read RNA/cDNA sequencing of TERT revealed 45 TERT mRNA variants including 13 known and 32 novel variants. Among the variants, TERT Delta 2-4, which lacks exons 2-4 but retains the original open reading frame, was selected for further study. Induced pluripotent stem cells and cancer cells express higher levels of TERT Delta 2-4 compared to primary human bronchial epithelial cells. Overexpression of TERT Delta 2-4 enhanced clonogenicity and resistance to cisplatin-induced cell death. Knockdown of endogenous TERT Delta 2-4 in Calu-6 cells reduced clonogenicity and resistance to cisplatin. Our results suggest that TERT Delta 2-4 enhances cancer cells' resistance to cell death. RNA sequencing following knockdown of Delta 2-4 TERT indicates that translation is downregulated and that mitochondrial related proteins are upregulated compared to controls. Overall, our data indicate that TERT produces many isoforms that influence the function of TERT and the abundance and activity of telomerase.
Keywords: Alternative RNA splicing; Lung cancer; TERT; Telomerase.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Kras mutations increase telomerase activity and targeting telomerase is a promising therapeutic strategy for Kras-mutant NSCLC.Oncotarget. 2017 Jan 3;8(1):179-190. doi: 10.18632/oncotarget.10162. Oncotarget. 2017. PMID: 27329725 Free PMC article.
-
Medaka tert produces multiple variants with differential expression during differentiation in vitro and in vivo.Int J Biol Sci. 2011 Apr 15;7(4):426-39. doi: 10.7150/ijbs.7.426. Int J Biol Sci. 2011. PMID: 21547060 Free PMC article.
-
Dynamics of TERT regulation via alternative splicing in stem cells and cancer cells.PLoS One. 2023 Aug 2;18(8):e0289327. doi: 10.1371/journal.pone.0289327. eCollection 2023. PLoS One. 2023. PMID: 37531400 Free PMC article.
-
The role of telomeres and telomerase reverse transcriptase isoforms in pluripotency induction and maintenance.RNA Biol. 2016 Aug 2;13(8):707-19. doi: 10.1080/15476286.2015.1134413. Epub 2016 Jan 19. RNA Biol. 2016. PMID: 26786236 Free PMC article. Review.
-
Physiological and pathological significance of human telomerase reverse transcriptase splice variants.Biochimie. 2013 Nov;95(11):1965-70. doi: 10.1016/j.biochi.2013.07.031. Epub 2013 Aug 9. Biochimie. 2013. PMID: 23933091 Review.
References
-
- Cohen, S. B. et al. Protein composition of catalytically active human telomerase from immortal cells. Science315, 1850–1853 (2007). - PubMed
-
- Greider, C. W. & Blackburn, E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell43, 405–413 (1985). - PubMed
-
- Santos, J. H., Meyer, J. N., Skorvaga, M., Annab, L. A. & Van Houten, B. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell3, 399–411 (2004). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

