S100P is a core gene for diagnosing and predicting the prognosis of sepsis
- PMID: 40000745
- PMCID: PMC11861684
- DOI: 10.1038/s41598-025-90858-8
S100P is a core gene for diagnosing and predicting the prognosis of sepsis
Abstract
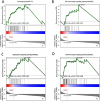
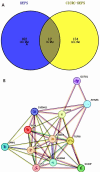
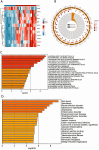
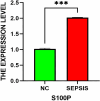
Sepsis, characterized as a severe systemic inflammatory response syndrome, typically originates from an exaggerated immune response to infection that gives rise to organ dysfunction. Serving as one of the predominant causes of death among critically ill patients, it's pressing to acquire an in-depth understanding of its intricate pathological mechanisms to strengthen diagnostic and therapeutic strategies. By integrating genomic, transcriptomic, proteomic, and metabolomic data across multiple biological levels, multi-omics research analysis has emerged as a crucial tool for unveiling the complex interactions within biological systems and unraveling disease mechanisms in recent years. Samples were collected from 23 cases of sepsis patients and 10 healthy volunteers from January 2019 to December 2020. The protein components in the samples were explored by independent data acquisition (DIA) analysis method, while Circular RNA (circRNA) categories were usually identified by RNA sequencing (RNA-seq) technology. Subsequent to the above steps, data quality monitoring was performed by employing software, and unqualified sequences were excluded, and conditions were set for differential expression network analysis (protein group and circRNA group were separately used log2 |FC|≥ 1 and log2 |FC|≥ 2, P < 0.050). Gene Ontology (GO) enrichment analysis and gene set enrichment analysis (GSEA) analysis were performed on common differentially expressed proteins, followed by protein-protein interaction between common differentially expressed genes and cytoscape software enrichment analysis, and subsequently its association with associated diseases (Disease Ontology (DO)) was investigated in an all-round manner. Afterwards, the distribution distinction of common differentially expressed genes in sepsis group and healthy volunteer group was displayed by heat map after Meta-analysis. Subsequent to the above procedures, pivotal targets with noticeable survival curve distinctions in two states were screened out after Meta-analysis. At last, their potential value was verified by in vitro cell experiment, which provided reference for further discussion of the diagnostic value and prognostic effect of target gene. A total of 174 DEPs and 308 DEcircRNAs were identified in the proteomics analysis, while a total of 12 common differentially expressed genes were identified after joint analysis. The protein-protein interaction (PPI) network suggested the degree of interaction between the dissimilar genes, and the heat map demonstrated their specific distribution in distinct groups. Through enrichment analysis, these proteins predominantly participated in a sequence of crucial processes such as intracellular material synthesis and secretion, changes in inflammatory receptors and immune inflammatory response. The meta-analysis identified that S100P is highly expressed in sepsis. As illustrated by the ROC curve, this gene has high clinical diagnostic value, and utimately confirmed its expression in sepsis through in vitro cell experiments. In these two groups of healthy people and septic patients, S100P demonstrated a more obvious trend of differential expression; Cell experiments also proved its value in diagnosis and prognosis judgment in sepsis; As a result, they may become diagnostic and prognostic markers for sepsis in clinical practice.
Keywords: Proteomics; RNA sequencing; S100P; Sepsis; Survival analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: Authors state no competing interests. Consent for publication: Written informed consent for publication was obtained from all participants. Ethics approval and consent to participate: The study was conducted in strict accordance with the rules of the Declaration of Helsinki.The study protocol has been approved by the ethics committee of the Affiliated Hospital ofSouthwest Medical University (Ethical Approval No. ky2018029). The Registration NumberSouthwest Medical University (Ethical Approval No. ky2018029). The Registration Number was ChiCTR1900021261.was ChiCTR1900021261. Informed consent: Informed consent was obtained from all individuals included in this study..
Figures






Similar articles
-
CTSO and HLA-DQA1 as biomarkers in sepsis-associated ARDS: insights from RNA sequencing and immune infiltration analysis.BMC Infect Dis. 2025 Mar 7;25(1):326. doi: 10.1186/s12879-025-10726-8. BMC Infect Dis. 2025. PMID: 40055592 Free PMC article.
-
SPP1 is a plasma biomarker associated with the dia gnosis and prediction of prognosis in sepsis.Sci Rep. 2024 Nov 8;14(1):27205. doi: 10.1038/s41598-024-78420-4. Sci Rep. 2024. PMID: 39516332 Free PMC article.
-
Exploring the prognostic and diagnostic value of lactylation-related genes in sepsis.Sci Rep. 2024 Oct 4;14(1):23130. doi: 10.1038/s41598-024-74040-0. Sci Rep. 2024. PMID: 39367086 Free PMC article.
-
Proteomics Combined with RNA Sequencing to Screen Biomarkers of Sepsis.Infect Drug Resist. 2022 Sep 21;15:5575-5587. doi: 10.2147/IDR.S380137. eCollection 2022. Infect Drug Resist. 2022. PMID: 36172619 Free PMC article.
-
The Diagnostic Utility of Host RNA Biosignatures in Adult Patients With Sepsis: A Systematic Review and Meta-Analysis.Crit Care Explor. 2025 Jan 31;7(2):e1212. doi: 10.1097/CCE.0000000000001212. eCollection 2025 Feb 1. Crit Care Explor. 2025. PMID: 39888601 Free PMC article.
References
-
- Atterton, B. et al. Sepsis associated delirium. Med. Lith.10.3390/medicina56050240 (2020).
-
- Grebenchikov, O. A. & Kuzovlev, A. N. Long-term outcomes after sepsis. Biochem. Mosc.86(5), 563–567. 10.1134/S0006297921050059 (2021). - PubMed
-
- Stephen, A. H., Montoya, R. L. & Aluisio, A. R. Sepsis and septic shock in low- and middle-income countries. Surg. Infect.21(7), 571–578. 10.1089/sur.2020.047 (2020). - PubMed
-
- Shi, X., Tan, S. & Tan, S. NLRP3 inflammasome in sepsis (review). Mol. Med. Rep.10.3892/mmr.2021.12153 (2021). - PubMed
-
- Opal, S. M. & Wittebole, X. Biomarkers of infection and sepsis. Crit. Care Clin.36(1), 11–22. 10.1016/j.ccc.2019.08.002 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

