Single-cell multi-omics analysis identifies SPP1+ macrophages as key drivers of ferroptosis-mediated fibrosis in ligamentum flavum hypertrophy
- PMID: 40001138
- PMCID: PMC11863437
- DOI: 10.1186/s40364-025-00746-6
Single-cell multi-omics analysis identifies SPP1+ macrophages as key drivers of ferroptosis-mediated fibrosis in ligamentum flavum hypertrophy
Abstract
Background: Ligamentum flavum hypertrophy (LFH) is a primary contributor to lumbar spinal stenosis. However, a thorough understanding of the cellular and molecular mechanisms driving LFH fibrotic progression remains incomplete.
Methods: Single-cell RNA sequencing (scRNA-seq) was performed to construct the single-cell map of human ligamentum flavum (LF) samples. An integrated multi-omics approach, encompassing scRNA-seq, bulk RNA sequencing (bulk RNA-seq), and Mendelian randomization (MR), was applied to conduct comprehensive functional analysis. Clinical tissue specimens and animal models were employed to further confirm the multi-omics findings.
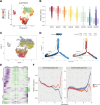
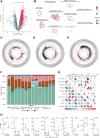
Results: ScRNA-seq provided a single-cell level view of the fibrotic microenvironment in LF, revealing significantly increased proportions of fibroblasts, myofibroblasts, and macrophages in LFH. Using transmission electron microscopy, single-cell gene set scoring, and MR analysis, ferroptosis was identified as a critical risk factor and pathway within LFH. Subcluster analysis of fibroblasts revealed functional heterogeneity among distinct subpopulations, highlighting the functional characteristics and the metabolic dynamics of fibroblast with a high ferroptosis score (High Ferro-score FB). The quantification of gene expression at single-cell level revealed that ferroptosis increased along with fibrosis in LFH specimens, a finding further validated in both human and mice tissue sections. Consistently, bulk RNA-seq confirmed increased proportions of fibroblasts and macrophages in LFH specimens, underscoring a strong correlation between these cell types through Spearman correlation analysis. Notably, subcluster analysis of the mononuclear phagocytes identified a specific subset of SPP1+ macrophages (SPP1+ Mac) enriched in LFH, which exhibited activation of fibrosis and ferroptosis-related metabolic pathways. Cell-cell communication analysis highlighted that SPP1+ Mac exhibited the strongest outgoing and incoming interactions among mononuclear phagocytes in the LFH microenvironment. Ligand-receptor analysis further revealed that the SPP1-CD44 axis could serve as a key mediator regulating the activity of High Ferro-score FB. Multiplex immunofluorescence confirmed substantial Collagen I deposition and reduced Ferritin Light Chain expression in regions with SPP1-CD44 co-localization in LFH specimens.
Conclusions: Our findings indicated that SPP1+ Mac may contribute to LFH fibrosis by regulating ferroptosis in High Ferro-score FB through the SPP1-CD44 axis. This study enhances our understanding of the cellular and molecular mechanisms underlying LFH progression, potentially improving early diagnostic strategies and identifying new therapeutic targets.
Keywords: Ferroptosis; Fibrosis; Ligamentum flavum hypertrophy; Macrophages.; Multi-omics analysis; Single-cell RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The collection and analysis of human LF specimen were approved by the Medical Ethics committee of Nanfang Hospital of Southern Medical University (Approval No: NFEC-2022-175). Written informed consent was obtained from all of the patients for being included in the study before tissue donation. The animal experiments followed the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) and were approved by the Animal Experiment Ethics Committee of Nanfang Hospital of Southern Medical University (Approval No: nfyy-2021-1021). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures







References
-
- Xiang Q, Zhao Y, Lin J, Jiang S, Li W. Epigenetic modifications in spinal ligament aging. Ageing Res Rev. 2022;77:101598. - PubMed
-
- Katz JN, Zimmerman ZE, Mass H, Makhni MC. Diagnosis and management of lumbar spinal stenosis: a review. JAMA. 2022;327:1688–99. - PubMed
-
- Fehlings MG, Tetreault L, Nater A, Choma T, Harrop J, Mroz T, et al. The aging of the global population: the changing epidemiology of disease and spinal disorders. Neurosurgery. 2015;77:S1–5. - PubMed
-
- Sun C, Zhang H, Wang X, Liu X. Ligamentum Flavum fibrosis and hypertrophy: molecular pathways, cellular mechanisms, and future directions. FASEB J. 2020;34:9854–68. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

