Classical Swine Fever Virus Envelope Glycoproteins Erns, E1, and E2 Activate IL-10-STAT1-MX1/OAS1 Antiviral Pathway via Replacing Classical IFNα/β
- PMID: 40001503
- PMCID: PMC11853677
- DOI: 10.3390/biom15020200
Classical Swine Fever Virus Envelope Glycoproteins Erns, E1, and E2 Activate IL-10-STAT1-MX1/OAS1 Antiviral Pathway via Replacing Classical IFNα/β
Abstract
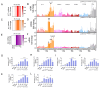
Classical swine fever (CSF) is an acute and often fatal disease caused by CSF virus (CSFV) infection. In the present study, we investigated the transcriptional profiles of peripheral blood mononuclear cells (PBMCs) in pigs infected with CSFV. The results revealed that CSFV inhibits IFNα/β production, but up-regulates the expression of signal transducer and activator of transcription 1 (STAT1); this result was verified in vitro. Interestingly, STAT1 is typically a downstream target of IFNα/β, raising the question of how CSFV can inhibit IFNα/β expression, yet up-regulate STAT1 expression. To explore this further, we observed that UV-treated CSFV induced STAT1 expression. Our results demonstrated that CSFV Erns, E1, and E2 could up-regulate STAT1 expression within the host cell cytoplasm and facilitate its translocation into the nucleus. The Erns, E1, and E2 proteins also separately induced the up-regulation of interleukin (IL)-10; IL-10 acts as the communicator connecting Erns, E1, and E2 proteins to STAT1, leading to the subsequent up-regulation, phosphorylation, and nuclear translocation of STAT1. Silencing of IL-10 down-regulated STAT1 expression. Finally, MX1 and OAS1 were identified as downstream targets of the IL-10-STAT1 pathway. In summary, a novel IL-10-STAT1 pathway independent of IFNα/β induced by CSFV Erns, E1, and E2 was identified in this study.
Keywords: CSFV; MX1; OAS1; STAT1; interleukin-10; surface glycoproteins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Interferon-Inducible Oligoadenylate Synthetase-Like Protein Acts as an Antiviral Effector against Classical Swine Fever Virus via the MDA5-Mediated Type I Interferon-Signaling Pathway.J Virol. 2017 May 12;91(11):e01514-16. doi: 10.1128/JVI.01514-16. Print 2017 Jun 1. J Virol. 2017. PMID: 28331099 Free PMC article.
-
P108 and T109 on E2 Glycoprotein Domain I Are Critical for the Adaptation of Classical Swine Fever Virus to Rabbits but Not for Virulence in Pigs.J Virol. 2020 Aug 17;94(17):e01104-20. doi: 10.1128/JVI.01104-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581110 Free PMC article.
-
Guanylate-Binding Protein 1, an Interferon-Induced GTPase, Exerts an Antiviral Activity against Classical Swine Fever Virus Depending on Its GTPase Activity.J Virol. 2016 Apr 14;90(9):4412-4426. doi: 10.1128/JVI.02718-15. Print 2016 May. J Virol. 2016. PMID: 26889038 Free PMC article.
-
Effects of early pregnancy on expression of interferon-stimulated gene 15, STAT1, OAS1, MX1, and IP-10 in ovine liver.Anim Sci J. 2020 Jan-Dec;91(1):e13378. doi: 10.1111/asj.13378. Anim Sci J. 2020. PMID: 32329195
-
Laboratory diagnosis, epizootiology, and efficacy of marker vaccines in classical swine fever: a review.Vet Q. 2000 Oct;22(4):182-8. doi: 10.1080/01652176.2000.9695054. Vet Q. 2000. PMID: 11087126 Review.
Cited by
-
Sow reproductive disorders: a key issue affecting the pig industry.Front Vet Sci. 2025 Mar 5;12:1535719. doi: 10.3389/fvets.2025.1535719. eCollection 2025. Front Vet Sci. 2025. PMID: 40110434 Free PMC article. Review.
References
-
- Postel A., Moennig V., Becher P. Classical swine fever in Europe--the current situation. Berl. Munch. Tierarztl. Wochenschr. 2014;126:468–475. - PubMed
MeSH terms
Substances
Grants and funding
- grant nos. 2021GXNSFAA220007 and 2017GXNSFAA198137/General Program of the Natural Science Foundation of Guangxi Province
- grant nos. 32360869, 31660722, and 31460653/the National Natural Science Foundation of China
- grant no. 2018GXNSFBA281170/the Youth Fund Project of the Natural Science Foundation of Guangxi Province
- grant no. 2023GXNSFDA026043/the Major Project of the Natural Science Foundation of Guangxi Province
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

