Chromosome-Level Genome Assembly of the Meishan Pig and Insights into Its Domestication Mechanisms
- PMID: 40003085
- PMCID: PMC11851914
- DOI: 10.3390/ani15040603
Chromosome-Level Genome Assembly of the Meishan Pig and Insights into Its Domestication Mechanisms
Abstract
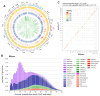
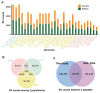
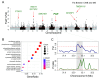
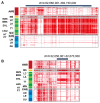
Pigs are essential agricultural animals, and among the various breeds, the Meishan pig, a native breed of China, is renowned for its high reproductive performance. This breed has been introduced to many countries to enhance local pig breeding programs. However, there have been limited genomic and population genetics studies focusing on Meishan pigs. We created a chromosomal-level genomic assembly using high-depth PacBio sequencing and Illumina sequencing data collected from a Meishan pig. Additionally, we analyzed whole-genome sequencing (WGS) data from Chinese boars and Meishan pigs to identify domestication selection signals within the Meishan breed. The assembled genome of the Meishan pig (MSjxau) was found to be 2.45 Gb in size, with a scaffold length of 139.17 Mb. The quality value was 37.06, and the BUSCO score was 96.2%, indicating good completeness, continuity, and accuracy. We annotated transposable elements, segmental duplication, and genes in the MSjxau genome. By combining these data with 28 publicly available genomes, we provide a high-quality structural variants resource for pigs. Furthermore, we identified 716 selective sweep intervals between Chinese wild pigs and Meishan pigs, where the selected gene PGR may be linked to the high fertility observed in Meishan pigs. Our study offers valuable genomic and variation resources for pig breeding and identifies several genes associated with the domestication of the Meishan pig. This lays the groundwork for further investigation into the genetic mechanisms behind complex traits in pigs.
Keywords: PacBio sequencing; domestication region; high fecundity; meishan pig genome; structural variation.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The Meishan pig genome reveals structural variation-mediated gene expression and phenotypic divergence underlying Asian pig domestication.Mol Ecol Resour. 2021 Aug;21(6):2077-2092. doi: 10.1111/1755-0998.13396. Epub 2021 Apr 26. Mol Ecol Resour. 2021. PMID: 33825319
-
Evidence of evolutionary history and selective sweeps in the genome of Meishan pig reveals its genetic and phenotypic characterization.Gigascience. 2018 May 1;7(5):giy058. doi: 10.1093/gigascience/giy058. Gigascience. 2018. PMID: 29790964 Free PMC article.
-
The selected genes NR6A1, RSAD2-CMPK2, and COL3A1 contribute to body size variation in Meishan pigs through different patterns.J Anim Sci. 2023 Jan 3;101:skad304. doi: 10.1093/jas/skad304. J Anim Sci. 2023. PMID: 37703114 Free PMC article.
-
CNV analysis of Meishan pig by next-generation sequencing and effects of AHR gene CNV on pig reproductive traits.J Anim Sci Biotechnol. 2020 Apr 21;11:42. doi: 10.1186/s40104-020-00442-5. eCollection 2020. J Anim Sci Biotechnol. 2020. PMID: 32337028 Free PMC article.
-
Uterine function in Meishan pigs.J Reprod Fertil Suppl. 1993;48:279-89. J Reprod Fertil Suppl. 1993. PMID: 8145211 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

