Bioinformatic Selection of Mannose-Specific Lectins from Allium genus as SARS-CoV-2 Inhibitors Analysing Protein-Protein Interaction
- PMID: 40003571
- PMCID: PMC11856470
- DOI: 10.3390/life15020162
Bioinformatic Selection of Mannose-Specific Lectins from Allium genus as SARS-CoV-2 Inhibitors Analysing Protein-Protein Interaction
Abstract
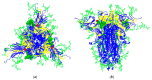
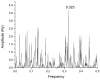
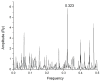
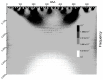
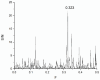
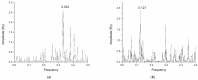
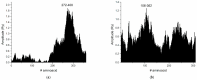
Mannose-specific lectins are carbohydrate-binding proteins known for their antiviral potential. This study uses a bioinformatic approach to investigate the possibility of lectins from Allium sativum (garlic) and Allium ursinum (wild garlic) as inhibitors of SARS-CoV-2 entry. The information spectrum method (ISM) identified key interaction frequencies between the SARS-CoV-2 spike protein and these lectins, explicitly targeting the receptor-binding domain (RBD) and glycosylated asparagine residues, including N234. Lectins from Allium species showed a high affinity for oligomannose-type glycans on the spike protein, potentially blocking virus entry by preventing the spike-ACE2 receptor interaction. We propose that Allium lectins are promising candidates for further experimental validation as SARS-CoV-2 inhibitors, offering potential therapeutic applications in managing viral infections.
Keywords: SARS-CoV-2; bioinformatics; informational spectrum method; lectins; spike protein.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration.Elife. 2020 Oct 26;9:e61552. doi: 10.7554/eLife.61552. Elife. 2020. PMID: 33103998 Free PMC article.
-
Man-Specific Lectins from Plants, Fungi, Algae and Cyanobacteria, as Potential Blockers for SARS-CoV, MERS-CoV and SARS-CoV-2 (COVID-19) Coronaviruses: Biomedical Perspectives.Cells. 2021 Jun 28;10(7):1619. doi: 10.3390/cells10071619. Cells. 2021. PMID: 34203435 Free PMC article. Review.
-
New mannose-specific lectins from garlic (Allium sativum) and ramsons (Allium ursinum) bulbs.Carbohydr Res. 1992 May 22;229(2):347-53. doi: 10.1016/s0008-6215(00)90580-9. Carbohydr Res. 1992. PMID: 1394291
-
Isolation and characterization of lectins and lectin-alliinase complexes from bulbs of garlic (Allium sativum) and ramsons (Allium ursinum).Glycoconj J. 1997 Apr;14(3):331-43. doi: 10.1023/a:1018570628180. Glycoconj J. 1997. PMID: 9147057
-
Plant lectins as versatile tools to fight coronavirus outbreaks.Glycoconj J. 2023 Feb;40(1):109-118. doi: 10.1007/s10719-022-10094-4. Epub 2022 Nov 24. Glycoconj J. 2023. PMID: 36418811 Free PMC article. Review.
Cited by
-
Nano-Phytomedicine: Harnessing Plant-Derived Phytochemicals in Nanocarriers for Targeted Human Health Applications.Molecules. 2025 Jul 29;30(15):3177. doi: 10.3390/molecules30153177. Molecules. 2025. PMID: 40807355 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

